Chitosan Rate of Uptake in HEK293 Cells is Influenced by Soluble versus Microparticle State and Enhanced by Serum-Induced Cell Metabolism and Lactate-Based Media Acidification
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Characterization of RITC-Chitosan and RITC-Chitosan Microparticles
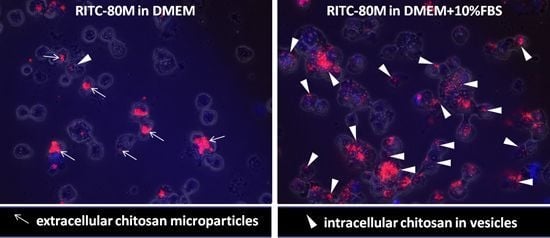
2.2. Serum Stimulates RITC-Chitosan Microparticle Uptake
2.3. Role of Serum and Lactate in Mediating Chitosan Uptake by HEK293 Cells
3. Experimental
3.1. Materials
3.2. Photoacoustic Fourrier Transform Infrared Spectroscopy (PAS FT-IR)
3.3. Particle Size and Zeta Potential
3.4. Measurement of RITC-Chitosan Solubility in Cell Culture Media
3.5. Flow Cytometry
3.6. Live Confocal Microscopy
3.7. Lactate and Glucose Measurements
3.8. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Shigemasa, Y.; Minami, S. Applications of chitin and chitosan for biomaterials. Biotechnol. Genet. Eng. Rev. 1996, 13, 383–420. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Reist, M.; Mayer, J.M.; Felt, O.; Gurny, R. Structure and interactions in chitosan hydrogels formed by complexation or aggregation for biomedical applications. Eur. J. Pharm. Biopharm. 2004, 57, 35–52. [Google Scholar] [CrossRef]
- Filion, D.; Lavertu, M.; Buschmann, M.D. Ionization and solubility of chitosan solutions related to thermosensitive chitosan/glycerol-phosphate systems. Biomacromolecules 2007, 8, 3224–3234. [Google Scholar] [CrossRef] [PubMed]
- Onishi, H.; Machida, Y. Biodegradation and distribution of water-soluble chitosan in mice. Biomaterials 1999, 20, 175–182. [Google Scholar] [CrossRef]
- Lin, C.W.; Lin, J.C. Characterization and blood coagulation evaluation of the water-soluble chitooligosaccharides prepared by a facile fractionation method. Biomacromolecules 2003, 4, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Ohara, N.; Hayashi, Y.; Yamada, S.; Kim, S.K.; Matsunaga, T.; Yanagiguchi, K.; Ikeda, T. Early gene expression analyzed by cDNA microarray and RT-PCR in osteoblasts cultured with water-soluble and low molecular chitooligosaccharide. Biomaterials 2004, 25, 1749–1754. [Google Scholar] [CrossRef] [PubMed]
- Varum, K.M.; Holme, H.K.; Izume, M.; Stokke, B.T.; Smidsrod, O. Determination of enzymatic hydrolysis specificity of partially N-acetylated chitosans. BBA-Gen. Subjects 1996, 1291, 5–15. [Google Scholar] [CrossRef]
- Tomihata, K.; Ikada, Y. In vitro and in vivo degradation of films of chitin and its deacetylated derivatives. Biomaterials 1997, 18, 567–575. [Google Scholar] [CrossRef]
- Huang, M.; Khor, E.; Lim, L.Y. Uptake and cytotoxicity of chitosan molecules and nanoparticles: Effects of molecular weight and degree of deacetylation. Pharm. Res. 2004, 21, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Okamoto, Y.; Takayama, T.; Shigemasa, Y.; Minami, S. Effect of N-acetyl-D-glucosamine and D-glucosamine oligomers on canine polymorphonuclear cells in vitro. Carbohydr. Polym. 1998, 36, 137–141. [Google Scholar] [CrossRef]
- Chevrier, A.; Hoemann, C.D.; Sun, J.; Buschmann, M.D. Chitosan-glycerol phosphate/blood implants increase cell recruitment, transient vascularization and subchondral bone remodeling in drilled cartilage defects. Osteoarthr. Cartil. 2007, 15, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Hoemann, C.D.; Chen, G.; Marchand, C.; Sun, J.; Tran-Khanh, N.; Chevrier, A.; Thibault, M.; Fernandes, M.G.J.; Poubelle, P.E.; Shive, M.S.; et al. Scaffold-guided subchondral bone repair: Implication of neutrophils and alternatively activated arginase-1+ macrophages. Am. J. Sports Med. 2010, 38, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Simard, P.; Galarneau, H.; Marois, S.; Rusu, D.; Hoemann, C.; Poubelle, P.; El-Gabalawy, H.; Fernandes, M. Neutrophils exhibit distinct phenotypes toward chitosans with different degrees of deacetylation: implications for cartilage repair. Arthritis Res. Ther. 2009, 11. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.Y.; Jang, M.K.; Nah, J.W. Influence of molecular weight on oral absorption of water soluble chitosans. J. Control. Release 2005, 102, 383–394. [Google Scholar] [CrossRef] [PubMed]
- de Campos, A.M.; Diebold, Y.; Carvalho, E.L.S.; Sanchez, A.; Alonso, M.J. Chitosan nanoparticles as new ocular drug delivery systems: In vitro stability, in vivo fate, and cellular toxicity. Pharm. Res. 2004, 21, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Casettari, L.; Vllasaliu, D.; Castagnino, E.; Stolnik, S.; Howdle, S.; Illum, L. PEGylated chitosan derivatives: Synthesis, characterizations and pharmaceutical applications. Prog. Polym. Sci. 2012, 37, 659–685. [Google Scholar] [CrossRef]
- Malette, W.G.; Quigley, H.J.; Gaines, R.D.; Johnson, N.D.; Rainer, W.G. Chitosan: A new hemostatic. Ann. Thorac. Surg. 1983, 36, 55–58. [Google Scholar] [CrossRef]
- Hoemann, C.D.; Hurtig, M.; Rossomacha, E.; Sun, J.; Chevrier, A.; Shive, M.S.; Buschmann, M.D. Chitosan-glycerol phosphate/blood implants improve hyaline cartilage repair in ovine microfracture defects. J. Bone Joint Surg.-Am. Vol. 2005, 87A, 2671–2686. [Google Scholar] [CrossRef] [PubMed]
- Onishi, H. Chitosan microparticles. J. Drug Deliv. Sci. Technol. 2010, 20, 15–22. [Google Scholar] [CrossRef]
- Hoemann, C.D.; Sun, J.; McKee, M.D.; Chevrier, A.; Rossomacha, E.; Rivard, G.E.; Hurtig, M.; Buschmann, M.D. Chitosan-glycerol phosphate/blood implants elicit hyaline cartilage repair integrated with porous subchondral bone in microdrilled rabbit defects. Osteoarthr. Cartil. 2007, 15, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Lafantaisie-Favreau, C.-H.; Guzman-Morales, J.; Sun, J.; Chen, G.; Harris, A.; Smith, T.D.; Carli, A.; Henderson, J.; Stanish, W.D.; Hoemann, C.D. Subchondral pre-solidified chitosan/blood implants elicit reproducible early osteochondral wound-repair responses including neutrophil and stromal cell chemotaxis, bone resorption and repair, enhanced repair tissue integration and delayed matrix deposition. BMC Musculoskelet. Disord. 2013, in press. [Google Scholar] [CrossRef] [PubMed]
- Thibault, M.; Astolfi, M.; Tran-Khanh, N.; Lavertu, M.; Darras, V.; Merzouki, A.; Buschmann, M.D. Excess polycation mediates efficient chitosan-based gene transfer by promoting lysosomal release of the polyplexes. Biomaterials 2011, 32, 4639–4646. [Google Scholar] [CrossRef] [PubMed]
- Lavertu, M.; Methot, S.; Tran-Khanh, N.; Buschmann, M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 2006, 27, 4815–4824. [Google Scholar] [CrossRef] [PubMed]
- Jean, M.; Smaoui, F.; Lavertu, M.; Methot, S.; Bouhdoud, L.; Buschmann, M.D.; Merzouki, A. Chitosan-plasmid nanoparticle formulations for IM and SC delivery of recombinant FGF-2 and PDGF-BB or generation of antibodies. Gene Ther. 2009, 16, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Ishii, T.; Okahata, Y. In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials 2001, 22, 2075–2080. [Google Scholar] [CrossRef]
- Nimesh, S.; Thibault, M.M.; Lavertu, M.; Buschmann, M.D. Enhanced Gene Delivery Mediated by Low Molecular Weight Chitosan/DNA Complexes: Effect of pH and Serum. Mol. Biotechnol. 2010, 46, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Morales, J.; El-Gabalawy, H.; Hoang, M.P.; Tran-Khanh, N.; McKee, M.D.; Wu, W.; Centola, M.; Hoemann, C.D. Effect of chitosan particles and dexamethasone on human bone marrow stromal cell osteogenesis and angiogenic factor secretion. Bone 2009, 45, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Morales, J.; Ariganello, M.B.; Hammami, I.; Thibault, M.; Jolicoeur, M.; Hoemann, C.D. Biodegradable chitosan particles induce chemokine release and negligible arginase-1 activity compared to IL-4 in murine bone marrow-derived macrophages. Biochem. Biophys. Res. Commun. 2011, 405, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Al Kobiasi, M.; Chua, B.Y.; Tonkin, D.; Jackson, D.C.; Mainwaring, D.E. Control of size dispersity of chitosan biopolymer microparticles and nanoparticles to influence vaccine trafficking and cell uptake. J. Biomed. Mater. Res. A 2012, 100A, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Nakamura, F.; Murakami, M.; Okumura, M.; Kadosawa, T.; Fujinag, T. Evaluation effects of chitosan for the extracellular matrix production by fibroblasts and the growth factors production by macrophages. Biomaterials 2001, 22, 2125–2130. [Google Scholar] [CrossRef]
- Usami, Y.; Okamoto, Y.; Minami, S.; Matsuhashi, A.; Kumazawa, N.H.; Tanioka, S.; Shigemasa, Y. Migration of canine neutrophils to chitin and chitosan. J. Vet. Med. Sci. 1994, 56, 1215–1216. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Ma, Z.S.; Khor, E.; Lim, L.Y. Uptake of FITC-chitosan nanoparticles by A549 cells. Pharm. Res. 2002, 19, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.S.; Lim, L.Y. Uptake of chitosan and associated insulin in Caco-2 cell monolayers: A comparison between chitosan molecules and chitosan nanoparticles. Pharm. Res. 2003, 20, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Ma, O.; Lavertu, M.; Sun, J.; Nguyen, S.; Buschmann, M.D.; Winnik, F.M.; Hoemann, C.D. Precise derivatization of structurally distinct chitosans with rhodamine B isothiocyanate. Carbohydr. Polym. 2008, 72, 616–624. [Google Scholar] [CrossRef]
- Benesch, J.; Tengvall, P. Blood protein adsorption onto chitosan. Biomaterials 2002, 23, 2561–2568. [Google Scholar] [CrossRef]
- Marchand, C.; Bachand, J.; Périnêt, J.; Baraghis, E.; Lamarre, M.; Rivard, G.E.; Crescenzo, G.D.; Hoemann, C.D. C3, C5, and factor B bind to chitosan without complement activation. J. Biomed. Mater. Res. A 2009, 93A, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Thibault, M.; Nimesh, S.; Lavertu, M.; Buschmann, M.D. Intracellular Trafficking and Decondensation Kinetics of Chitosan-pDNA Polyplexes. Mol. Ther. 2010, 18, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.L.; Ferreira, M.C.; Marvao, M.R.; Rocha, J. An optimised method to determine the degree of acetylation of chitin and chitosan by FTIR spectroscopy. Int. J. Biol. Macromol. 2002, 31, 1–8. [Google Scholar] [CrossRef]
- Cardenas, G.; Cabrera, G.; Taboada, E.; Miranda, S.P. Chitin characterization by SEM, FTIR, XRD, and C-13 cross polarization/mass angle spinning NMR. J. Appl. Polym. Sci. 2004, 93, 1876–1885. [Google Scholar] [CrossRef]
- Yohannes, G.; Wiedmer, S.K.; Elomaa, M.; Jussila, M.; Aseyev, V.; Riekkola, M.-L. Thermal aggregation of bovine serum albumin studied by asymmetrical flow field-flow fractionation. Anal. Chim. Acta 2010, 675, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, R.M.; Molitoris, B.A. Gentamicin traffics retrograde through the secretory pathway and is released in the cytosol via the endoplasmic reticulum. Am. J. Physiol. Renal Physiol. 2004, 286, F617–F624. [Google Scholar] [CrossRef] [PubMed]
- Gorochovceva, N.; Makuska, R. Synthesis and study of water-soluble chitosan-O-poly(ethylene glycol) graft copolymers. Eur. Polym. J. 2004, 40, 685–691. [Google Scholar] [CrossRef]
- Berger, M.; Hammer, C.H. Complement and Complement Reactions; Springer-Verlag: New York, NY, USA, 1988; pp. 4–17. [Google Scholar]
- Ayache, S.; Panelli, M.C.; Byrne, K.M.; Slezak, S.; Leitman, S.F.; Marincola, F.M.; Stroncek, D.F. Comparison of proteomic profiles of serum, plasma, and modified media supplements used for cell culture and expansion. J. Transl. Med. 2006, 4. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.C.; Lindsay, M.R.; Luetterforst, R.; Hamilton, N.; Simpson, F.; Parton, R.G.; Gleeson, P.A.; Teasdale, R.D. Visualisation of macropinosome maturation by the recruitment of sorting nexins. J. Cell Sci. 2006, 119, 3967–3980. [Google Scholar] [CrossRef] [PubMed]
- Koivusalo, M.; Welch, C.; Hayashi, H.; Scott, C.C.; Kim, M.; Alexander, T.; Touret, N.; Hahn, K.M.; Grinstein, S. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 2010, 188, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Ceriotti, L.; Kob, A.; Drechsler, S.; Ponti, J.; Thedinga, E.; Colpo, P.; Ehret, R.; Rossi, F. Online monitoring of BALB/3T3 metabolism and adhesion with multiparametric chip-based system. Anal. Biochem. 2007, 371, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Swanson, J.A.; Watts, C. Macropinocytosis. Trends Cell Biol. 1995, 5, 424–428. [Google Scholar] [CrossRef]
- Ehrenberg, M.S.; Friedman, A.E.; Finkelstein, J.N.; Oberdorster, G.; McGrath, J.L. The influence of protein adsorption on nanoparticle association with cultured endothelial cells. Biomaterials 2009, 30, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Greyner, H.J.; Wiraszka, T.; Zhang, L.S.; Petroll, W.M.; Mummert, M.E. Inducible macropinocytosis of hyaluronan in B16-F10 melanoma cells. Matrix Biol. 2010, 29, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Gratton, S.E.A.; Ropp, P.A.; Pohlhaus, P.D.; Luft, J.C.; Madden, V.J.; Napier, M.E.; DeSimone, J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 11613–11618. [Google Scholar] [CrossRef] [PubMed]
- Corsi, K.; Chellat, F.; Yahia, L.; Fernandes, J.C. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials 2003, 24, 1255–1264. [Google Scholar] [CrossRef]
- Lafantaisie-Favreau, C.-H.; Sun, J.; Chen, G.; Rusu, D.; Poubelle, P.; Hoemann, C.D. Implication of mast cells in chitosan/blood implant induction of in vivo chemotaxis and alternative macrophage activation. Presented at Canadian Connective Tissue Conference, Montreal, Canada, 27 May 2011. [Google Scholar]
- Kas, H.S. Chitosan: Properties, preparations and application to microparticulate systems. J. Microencapsul. 1997, 14, 689–711. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of RITC-95M and RITC-80M chitosan are available from the authors. |












| Method of insoluble chitosan microparticle preparation | Chitosans analyzed | Application |
|---|---|---|
| Neutral precipitation of acid-soluble chitosan in vivo or in vitro | ||
| Mix acid-soluble chitosan solution or chitosan-glycerol phosphate solution pH 4–6.8 with whole blood and/or directly apply to bleeding tissues | 80% DDA 75%–82% DDA, 150–250 kDa | Hemostatic in vivo [17] Bone and articular cartilage repair in vivo [20] |
| Inject mixtures of chitosan-DNA, with a molar excess of chitosan, in intramuscular, subcutaneous sites | 92% DDA, 10 kDa 80% DDA, 10 kDa 80% DDA, 80 kDa | Gene delivery in vivo [24] |
| Combine soluble chitosan at 5- to 10-fold molar excess with DNA, pipette into cell culture medium DMEM+10% serum pH 7.6 | 80% DDA, 15 kDa 92% DDA, >100 kDa 94% DDA, 52 kDa | In vitro DNA delivery: A549, Hela, B16 cells, HEK293 cells [25,26] |
| Pipette acid-soluble chitosan pH 5.0 into basal media pH 7.6 (DMEM, αMEM, RPMI±10% to 16% fetal bovine serum) | 80% DDA, 179 kDa 81% DDA, 35 kDa 80% DDA, 179 kDa 95% DDA, 168 kDa 92% DDA, 10 kDa | In vitro bone marrow stromal cell osteogenesis [27] In vitro macrophage activation [28] In vitro neutrophil chemotaxis, degranulation, chitosan uptake [14] In vitro chitosan-HEK293 cell adsorption/uptake [26] |
| Pre-formed chitosan microparticles | ||
| Pre-formed microparticles (glutaraldehyde and Tween surfactant) injected into the mouse foot pad or added to DMEM+heat-inactivated 10% fetal bovine serum | 75%–85% DDA, 164 kDa | Vaccine, lymph node trafficking in vivo [29] In vitro HEK293, A549, RAW264 cell uptake [29] |
| 1 µm or 3.5 µm pre-formed chitosan microparticles added to RPMI+10% serum, pH 7.2 | ≥80% DDA | Wound-repair applications: in vitro neutrophil chemotaxis [30] in vitro macrophage activation [31] |
| chitosan | Viscosity (mPa.s) ^ | Chitosan DDA (%) # | Mn (kDa) | PDI (Mw/Mn) | RITC/chitosan % mol/mol |
|---|---|---|---|---|---|
| RITC-95M * | 2,964 | 94.6 | 177 | 1.1 | 0.6 |
| RITC-95L * | 197 | 94.6 | 102 | 1.2 | 0.6 |
| RITC-80M * | 1,422 | 80.6 | 144 | 1.3 | 0.5 |
| RITC-80L * | 178 | 80.2 | 108 | 1.5 | 0.5 |
| RITC-82M ** | N.D. | 81.7 | 241 | 1.1 | 0.9 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hoemann, C.D.; Guzmán-Morales, J.; Tran-Khanh, N.; Lavallée, G.; Jolicoeur, M.; Lavertu, M. Chitosan Rate of Uptake in HEK293 Cells is Influenced by Soluble versus Microparticle State and Enhanced by Serum-Induced Cell Metabolism and Lactate-Based Media Acidification. Molecules 2013, 18, 1015-1035. https://doi.org/10.3390/molecules18011015
Hoemann CD, Guzmán-Morales J, Tran-Khanh N, Lavallée G, Jolicoeur M, Lavertu M. Chitosan Rate of Uptake in HEK293 Cells is Influenced by Soluble versus Microparticle State and Enhanced by Serum-Induced Cell Metabolism and Lactate-Based Media Acidification. Molecules. 2013; 18(1):1015-1035. https://doi.org/10.3390/molecules18011015
Chicago/Turabian StyleHoemann, Caroline D., Jessica Guzmán-Morales, Nicolas Tran-Khanh, Geneviève Lavallée, Mario Jolicoeur, and Marc Lavertu. 2013. "Chitosan Rate of Uptake in HEK293 Cells is Influenced by Soluble versus Microparticle State and Enhanced by Serum-Induced Cell Metabolism and Lactate-Based Media Acidification" Molecules 18, no. 1: 1015-1035. https://doi.org/10.3390/molecules18011015