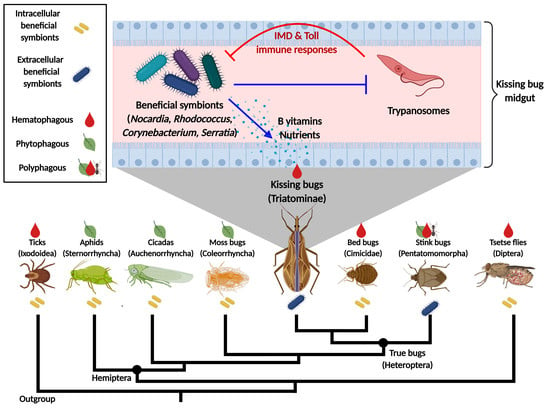
The Role of Bacterial Symbionts in Triatomines: An Evolutionary Perspective
Abstract
:1. Introduction
2. Insect–Microbe Symbioses
3. Evolution of Beneficial Symbioses in the Hemiptera
3.1. Sternorrhyncha and Auchenorrhyncha Symbioses
3.2. Heteroptera Symbioses
4. Triatomine Symbioses
4.1. Classical Studies with Rhodococcus rhodnii
4.2. Microbiome Studies
4.3. Effects of Trypanosomes on the Microbiota
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosenberg, E.; Zilber-Rosenberg, I. The hologenome concept of evolution after 10 years. Microbiome 2018, 6, 78. [Google Scholar] [CrossRef]
- Sudakaran, S.; Kost, C.; Kaltenpoth, M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017, 25, 375–390. [Google Scholar] [CrossRef]
- Gil, R.; Latorre, A. Unity makes strength: A review on mutualistic symbiosis in representative insect clades. Life 2019, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Janson, E.M.; Stireman, J.O.; Singer, M.S.; Abbot, P. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution 2008, 62, 997–1012. [Google Scholar] [CrossRef] [Green Version]
- Buchner, P. Endosymbiosis of Animals with Plant Microorganisms; Interscience Publishers/John Wiley: Hoboken, NJ, USA, 1965; p. 909. [Google Scholar]
- Zaidman-Rémy, A.; Vigneron, A.; Weiss, B.L.; Heddi, A. What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus. BMC Microbiol. 2018, 18, 150. [Google Scholar] [CrossRef] [Green Version]
- Wernegreen, J.J. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 2002, 3, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.J.; Moran, N.A. Links between metamorphosis and symbiosis in holometabolous insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20190068. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Ruano, S.M.; Škochová, V.; Rego, R.O.M.; Schmidt, J.O.; Roachell, W.; Hypša, V.; Nováková, E. Microbiomes of north american triatominae: The grounds for chagas disease epidemiology. Front. Microbiol. 2018, 9, 1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, P.R.; Paris, V.; Rolff, J. Immune gene regulation in the gut during metamorphosis in a holo- versus a hemimetabolous insect. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20190073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez-Moo, P.; Cruz-Rosales, M.; Ibarra-Laclette, E.; Desgarennes, D.; Huerta, C.; Lamelas, A. Diversity and Composition of the Gut Microbiota in the Developmental Stages of the Dung Beetle Copris incertus Say (Coleoptera, Scarabaeidae). Front. Microbiol. 2020, 11, 1698. [Google Scholar] [CrossRef]
- Majumder, R.; Sutcliffe, B.; Taylor, P.W.; Chapman, T.A. Microbiome of the Queensland Fruit Fly through Metamorphosis. Microorganisms 2020, 8, 795. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, P.R.; Rolff, J. Host and Symbiont Jointly Control Gut Microbiota during Complete Metamorphosis. PLoS Pathog. 2015, 11, e1005246. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Aksoy, S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 1999, 8, 125–132. [Google Scholar] [CrossRef]
- Stoll, S.; Feldhaar, H.; Fraunholz, M.J.; Gross, R. Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 2010, 10, 308. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, T.L.; Douglas, A.E. The impact of aposymbiosis on amino acid metabolism of pea aphids (Acyrthosiphon pisum). Ent. Exp. Appl. 1996, 80, 279–282. [Google Scholar] [CrossRef]
- Davey, K.G. The modes of action of juvenile hormones: Some questions we ought to ask. Insect Biochem. Mol. Biol. 2000, 30, 663–669. [Google Scholar] [CrossRef]
- Davey, K.G. Reproduction in the Insects, 2nd ed.; Oliver and Boyd: Edinburgh, Scotland, 1965; p. 96. [Google Scholar]
- Wigglesworth, V.B. The Principles of Insect Physiology; Springer: Dordrecht, The Netherlands, 1972. [Google Scholar]
- Pérez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Batra, L.R. Ecology of ambrosia fungi and their dissemination by beetles. Trans. Kans. Acad. Sci. 1963, 66, 213. [Google Scholar] [CrossRef]
- Maire, J.; Vincent-Monégat, C.; Balmand, S.; Vallier, A.; Hervé, M.; Masson, F.; Parisot, N.; Vigneron, A.; Anselme, C.; Perrin, J.; et al. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc. Natl. Acad. Sci. USA 2019, 116, 5623–5632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Login, F.H.; Balmand, S.; Vallier, A.; Vincent-Monégat, C.; Vigneron, A.; Weiss-Gayet, M.; Rochat, D.; Heddi, A. Antimicrobial peptides keep insect endosymbionts under control. Science 2011, 334, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Trappeniers, K.; Matetovici, I.; Van Den Abbeele, J.; De Vooght, L. The Tsetse Fly Displays an Attenuated Immune Response to Its Secondary Symbiont, Sodalis glossinidius. Front. Microbiol. 2019, 10, 1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Dos-Santos, A.L.A.; Huang, W.; Liu, K.C.; Oshaghi, M.A.; Wei, G.; Agre, P.; Jacobs-Lorena, M. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 2017, 357, 1399–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.C.C.; Duncan, R.P. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. USA 2015, 112, 10255–10261. [Google Scholar] [CrossRef] [Green Version]
- Sacchi, L.; Grigolo, A.; Biscaldi, G.; Laudani, U. Effects of heat treatment on the symbiotic system of Blattoidea: Morphofunctional alterations of bacteriocytes. Bolletino di Zoologia 1993, 60, 271–279. [Google Scholar] [CrossRef]
- Braendle, C.; Miura, T.; Bickel, R.; Shingleton, A.W.; Kambhampati, S.; Stern, D.L. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003, 1, E21. [Google Scholar] [CrossRef]
- Kuechler, S.M.; Fukatsu, T.; Matsuura, Y. Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 2019, 21, 4378–4394. [Google Scholar] [CrossRef]
- Hansen, A.K.; Moran, N.A. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA 2011, 108, 2849–2854. [Google Scholar] [CrossRef] [Green Version]
- Hongoh, Y. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol. Life Sci. 2011, 68, 1311–1325. [Google Scholar] [CrossRef]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Matsuura, Y.; Watanabe, H.; Fujishima, M.; Brune, A. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. USA 2018, 115, E11996–E12004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Li, H.; Chevrette, M.G.; Zhang, L.; Cao, L.; Zhou, H.; Zhou, X.; Zhou, Z.; Pope, P.B.; Currie, C.R.; et al. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J. 2019, 13, 104–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourguignon, T.; Lo, N.; Dietrich, C.; Šobotník, J.; Sidek, S.; Roisin, Y.; Brune, A.; Evans, T.A. Rampant host switching shaped the termite gut microbiome. Curr. Biol. 2018, 28, 649–654.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervé, V.; Liu, P.; Dietrich, C.; Sillam-Dussès, D.; Stiblik, P.; Šobotník, J.; Brune, A. Phylogenomic analysis of 589 metagenome-assembled genomes encompassing all major prokaryotic lineages from the gut of higher termites. PeerJ 2020, 8, e8614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scolari, F.; Casiraghi, M.; Bonizzoni, M. Aedes spp. and Their Microbiota: A Review. Front. Microbiol. 2019, 10, 2036. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.C.-N.; Chaston, J.M.; Douglas, A.E. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013, 7, 1922–1932. [Google Scholar] [CrossRef] [Green Version]
- Coon, K.L.; Brown, M.R.; Strand, M.R. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 2016, 25, 5806–5826. [Google Scholar] [CrossRef] [Green Version]
- Aksoy, E.; Telleria, E.L.; Echodu, R.; Wu, Y.; Okedi, L.M.; Weiss, B.L.; Aksoy, S.; Caccone, A. Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl. Environ. Microbiol. 2014, 80, 4301–4312. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. Requirement of pea aphids (Acyrthosiphon pisum) for their symbiotic bacteria. Entomol. Exp. Appl. 1992, 65, 195–198. [Google Scholar] [CrossRef]
- Schaub, G.A.; Eichler, S. The effects of aposymbiosis and of an infection with Blastocrithidia triatomae (Trypanosomatidae) on the tracheal system of the reduviid bugs Rhodnius prolixus and Triatoma infestans. J. Insect Physiol. 1998, 44, 131–140. [Google Scholar]
- Heys, C.; Lizé, A.; Blow, F.; White, L.; Darby, A.; Lewis, Z.J. The effect of gut microbiota elimination in Drosophila melanogaster: A how-to guide for host-microbiota studies. Ecol. Evol. 2018, 8, 4150–4161. [Google Scholar] [CrossRef] [PubMed]
- Bell, W.J.; Adiyodi, K.G. The American Cockroach, 1st ed.; Springer: Dordrecht, The Netherlands, 1982. [Google Scholar]
- Kikuchi, Y.; Hosokawa, T.; Nikoh, N.; Meng, X.-Y.; Kamagata, Y.; Fukatsu, T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009, 7, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brecher, G.; Wigglesworth, V.B. The transmission of Actinomyces rhodnii Erikson in Rhodnius prolixus stål (hemiptera) and its influence on the growth of the host. Parasitology 1944, 35, 220. [Google Scholar] [CrossRef]
- Hammer, T.J.; Sanders, J.G.; Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019, 366, fnz117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Cortés, J.G.; García-Contreras, R.; Bucio-Torres, M.I.; Cabrera-Bravo, M.; Córdoba-Aguilar, A.; Benelli, G.; Salazar-Schettino, P.M. Bacterial symbionts in human blood-feeding arthropods: Patterns, general mechanisms and effects of global ecological changes. Acta Tropica 2018, 186, 69–101. [Google Scholar] [CrossRef]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [Green Version]
- Brownlie, J.C.; Johnson, K.N. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009, 17, 348–354. [Google Scholar] [CrossRef]
- O’Neill, S.L.; Gooding, R.H.; Aksoy, S. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med. Vet. Entomol. 1993, 7, 377–383. [Google Scholar] [CrossRef]
- Hosokawa, T.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol. Ecol. 2005, 54, 471–477. [Google Scholar] [CrossRef]
- Hansen, A.K.; Trumble, J.T.; Stouthamer, R.; Paine, T.D. A new Huanglongbing Species, “Candidatus Liberibacter psyllaurous,” found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Appl. Environ. Microbiol. 2008, 74, 5862–5865. [Google Scholar] [CrossRef] [Green Version]
- Caspi-Fluger, A.; Inbar, M.; Mozes-Daube, N.; Katzir, N.; Portnoy, V.; Belausov, E.; Hunter, M.S.; Zchori-Fein, E. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. Biol. Sci. 2012, 279, 1791–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltenpoth, M.; Winter, S.A.; Kleinhammer, A. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 2009, 69, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigglesworth, V.B. Symbiotic Bacteria in a Blood-sucking Insect, Rhodnius Prolixus Stål. (Hemiptera, Triatomidae). Parasitology 1936, 28, 284–289. [Google Scholar] [CrossRef]
- Hosokawa, T.; Kikuchi, Y.; Shimada, M.; Fukatsu, T. Symbiont acquisition alters behaviour of stinkbug nymphs. Biol. Lett. 2008, 4, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, T.; Kikuchi, Y.; Nikoh, N.; Shimada, M.; Fukatsu, T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006, 4, e337. [Google Scholar] [CrossRef] [PubMed]
- Fukatsu, T.; Hosokawa, T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 2002, 68, 389–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, C.M.; Coffey, P.L.; DeLay, B.D.; Dively, G.P. The importance of gut symbionts in the development of the brown marmorated stink bug, Halyomorpha halys (Stål). PLoS ONE 2014, 9, e90312. [Google Scholar] [CrossRef]
- Bakula, M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Invertebr. Pathol. 1969, 14, 365–374. [Google Scholar] [CrossRef]
- Guégan, M.; Zouache, K.; Démichel, C.; Minard, G.; Tran Van, V.; Potier, P.; Mavingui, P.; Valiente Moro, C. The mosquito holobiont: Fresh insight into mosquito-microbiota interactions. Microbiome 2018, 6, 49. [Google Scholar] [CrossRef]
- Rolff, J.; Johnston, P.R.; Reynolds, S. Complete metamorphosis of insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20190063. [Google Scholar] [CrossRef]
- Tóth, E.M.; Hell, E.; Kovács, G.; Borsodi, A.K.; Márialigeti, K. Bacteria isolated from the different developmental stages and larval organs of the obligate parasitic fly, Wohlfahrtia magnifica (Diptera: Sarcophagidae). Microb. Ecol. 2006, 51, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Andongma, A.A.; Wan, L.; Dong, Y.-C.; Li, P.; Desneux, N.; White, J.A.; Niu, C.-Y. Pyrosequencing reveals a shift in symbiotic bacteria populations across life stages of Bactrocera dorsalis. Sci. Rep. 2015, 5, 9470. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Du, K.; Sun, C.; Vimalanathan, A.; Liang, X.; Li, Y.; Wang, B.; Lu, X.; Li, L.; Shao, Y. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef] [Green Version]
- Moll, R.M.; Romoser, W.S.; Modrzakowski, M.C.; Moncayo, A.C.; Lerdthusnee, K. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J. Med. Entomol. 2001, 38, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.J.; McMillan, W.O.; Fierer, N. Metamorphosis of a butterfly-associated bacterial community. PLoS ONE 2014, 9, e86995. [Google Scholar] [CrossRef]
- Arias-Cordero, E.; Ping, L.; Reichwald, K.; Delb, H.; Platzer, M.; Boland, W. Comparative evaluation of the gut microbiota associated with the below- and above-ground life stages (larvae and beetles) of the forest cockchafer, Melolontha hippocastani. PLoS ONE 2012, 7, e51557. [Google Scholar] [CrossRef] [Green Version]
- Mereghetti, V.; Chouaia, B.; Limonta, L.; Locatelli, D.P.; Montagna, M. Evidence for a conserved microbiota across the different developmental stages of Plodia interpunctella. Insect Sci. 2019, 26, 466–478. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Da Mota, F.F.; Marinho, L.P.; de Carvalho Moreira, C.J.; Lima, M.M.; Mello, C.B.; Garcia, E.S.; Carels, N.; Azambuja, P. Cultivation-independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Negl. Trop. Dis. 2012, 6, e1631. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.-H.; Roh, S.W.; Whon, T.W.; Jung, M.-J.; Kim, M.-S.; Park, D.-S.; Yoon, C.; Nam, Y.-D.; Kim, Y.-J.; Choi, J.-H.; et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 2014, 80, 5254–5264. [Google Scholar] [CrossRef] [Green Version]
- Onchuru, T.O.; Javier Martinez, A.; Ingham, C.S.; Kaltenpoth, M. Transmission of mutualistic bacteria in social and gregarious insects. Curr. Opin. Insect Sci. 2018, 28, 50–58. [Google Scholar] [CrossRef]
- Johnson, K.P.; Dietrich, C.H.; Friedrich, F.; Beutel, R.G.; Wipfler, B.; Peters, R.S.; Allen, J.M.; Petersen, M.; Donath, A.; Walden, K.K.O.; et al. Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. USA 2018, 115, 12775–12780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, C.W.; Panizzi, A.R. Heteroptera of Economic Importance, 1st ed.; Schaefer, C.W., Panizzi, A.R., Eds.; CRC Press: Boca Raton, CA, USA, 2000. [Google Scholar]
- Douglas, A.E.; van Emden, H.F. Nutrition and symbiosis. In Aphids as Crop Pests; van Emden, H.F., Harrington, R., Eds.; CABI: Wallingford, Oxforshire, UK, 2007; pp. 115–134. [Google Scholar]
- World Health Organization. Global Vector Control Response 2017–2030; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Wang, Y.-H.; Wu, H.-Y.; Rédei, D.; Xie, Q.; Chen, Y.; Chen, P.-P.; Dong, Z.-E.; Dang, K.; Damgaard, J.; Štys, P.; et al. When did the ancestor of true bugs become stinky? Disentangling the phylogenomics of Hemiptera-Heteroptera. Cladistics 2017, 35, 42–66. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cui, Y.; Rédei, D.; Baňař, P.; Xie, Q.; Štys, P.; Damgaard, J.; Chen, P.; Yi, W.; Wang, Y.; et al. Phylogenetic divergences of the true bugs (Insecta: Hemiptera: Heteroptera), with emphasis on the aquatic lineages: The last piece of the aquatic insect jigsaw originated in the Late Permian/Early Triassic. Cladistics 2016, 32, 390–405. [Google Scholar] [CrossRef]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Sandström, J.; Moran, N. How nutritionally imbalanced is phloem sap for aphids? Entomol. Exp. Appl. 1999, 91, 203–210. [Google Scholar] [CrossRef]
- Dolling, W.R. The Hemiptera; Oxford University Press: Oxford, UK, 1991. [Google Scholar]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef] [Green Version]
- Moran, N.A.; Munson, M.A.; Baumann, P.; Ishikawa, H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B. 1993, 253, 167–171. [Google Scholar]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef] [Green Version]
- Sabree, Z.L.; Degnan, P.H.; Moran, N.A. Chromosome stability and gene loss in cockroach endosymbionts. Appl. Environ. Microbiol. 2010, 76, 4076–4079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, G.M.; McCutcheon, J.P.; MacDonald, B.R.; Romanovicz, D.; Moran, N.A. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio. 2014, 5, e01697-14. [Google Scholar]
- Lamelas, A.; Gosalbes, M.J.; Manzano-Marín, A.; Peretó, J.; Moya, A.; Latorre, A. Serratia symbiotica from the aphid Cinara cedri: A missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011, 7, e1002357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, R.; Moran, N.A. Swapping symbionts in spittlebugs: Evolutionary replacement of a reduced genome symbiont. ISME J. 2014, 8, 1237–1246. [Google Scholar] [CrossRef] [Green Version]
- Manzano-Marín, A.; Simon, J.-C.; Latorre, A. Reinventing the Wheel and Making It Round Again: Evolutionary Convergence in Buchnera-Serratia Symbiotic Consortia between the Distantly Related Lachninae Aphids Tuberolachnus salignus and Cinara cedri. Genome Biol. Evol. 2016, 8, 1440–1458. [Google Scholar] [CrossRef] [Green Version]
- Meseguer, A.S.; Manzano-Marín, A.; Coeur d’Acier, A.; Clamens, A.L.; Godefroid, M.; Jousselin, E. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol. Ecol. 2017, 26, 2363–2378. [Google Scholar] [CrossRef]
- Lamelas, A.; Pérez-Brocal, V.; Gómez-Valero, L.; Gosalbes, M.J.; Moya, A.; Latorre, A. Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in aphid species of the subfamily lachninae. Appl. Environ. Microbiol. 2008, 74, 4236–4240. [Google Scholar] [CrossRef] [Green Version]
- Bell-Roberts, L.; Douglas, A.E.; Werner, G.D.A. Match and mismatch between dietary switches and microbial partners in plant sap-feeding insects. Proc. Biol. Sci. 2019, 286, 20190065. [Google Scholar] [CrossRef] [Green Version]
- Salem, H.; Florez, L.; Gerardo, N.; Kaltenpoth, M. An out-of-body experience: The extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. Biol. Sci. 2015, 282, 20142957. [Google Scholar] [CrossRef] [Green Version]
- Ben-Yakir, D. Growth retardation of Rhodnius prolixus symbionts by immunizing host against Nocardia (Rhodococcus) rhodnii. J. Insect Physiol. 1987, 33, 379–383. [Google Scholar] [CrossRef]
- Sudakaran, S.; Salem, H.; Kost, C.; Kaltenpoth, M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Mol. Ecol. 2012, 21, 6134–6151. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011, 5, 446–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasgow, H. The gastric cæca and the cæcal bacteria of the Heteroptera. Biol Bull. 1914, 26, 101–107. [Google Scholar] [CrossRef]
- Baines, S. The role of the symbiotic bacteria in the nutrition of Rhodnius prolixus (Hemiptera). J. Exp. Biol. 1956, 33, 533–541. [Google Scholar]
- Davis, G.R. Essential dietary amino acids for growth of larvae of the yellow mealworm, Tenebrio molitor L. J. Nutr. 1975, 105, 1071–1075. [Google Scholar] [CrossRef] [Green Version]
- Nakabachi, A.; Yamashita, A.; Toh, H.; Ishikawa, H.; Dunbar, H.E.; Moran, N.A.; Hattori, M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Sci. 2006, 314, 267. [Google Scholar] [CrossRef] [Green Version]
- Sloan, D.B.; Moran, N.A. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 2012, 8, 986–989. [Google Scholar] [CrossRef] [Green Version]
- Bennett, G.M.; Moran, N.A. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a Phloem-feeding insect. Genome Biol. Evol. 2013, 5, 1675–1688. [Google Scholar] [CrossRef] [Green Version]
- Latorre, A.; Manzano-Marín, A. Dissecting genome reduction and trait loss in insect endosymbionts. Ann. NY Acad. Sci. 2017, 1389, 52–75. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA 2009, 106, 15394–15399. [Google Scholar] [CrossRef] [Green Version]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef] [PubMed]
- International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313.
- Wang, J.; Wu, Y.; Yang, G.; Aksoy, S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. USA 2009, 106, 12133–12138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maire, J.; Vincent-Monégat, C.; Masson, F.; Zaidman-Rémy, A.; Heddi, A. An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp. Microbiome 2018, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Michalik, A.; Szklarzewicz, T.; Jankowska, W.; Wieczorek, K. Endosymbiotic microorganisms of aphids (Hemiptera: Sternorrhyncha: Aphidoidea): Ultrastructure, distribution and transovarial transmission. Eur. J. Entomol. 2014, 111, 91–104. [Google Scholar] [CrossRef] [Green Version]
- McCutcheon, J.P.; von Dohlen, C.D. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 2011, 21, 1366–1372. [Google Scholar] [CrossRef] [Green Version]
- Husnik, F.; Nikoh, N.; Koga, R.; Ross, L.; Duncan, R.P.; Fujie, M.; Tanaka, M.; Satoh, N.; Bachtrog, D.; Wilson, A.C.C.; et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 2013, 153, 1567–1578. [Google Scholar] [CrossRef] [Green Version]
- Rosenblueth, M.; Sayavedra, L.; Sámano-Sánchez, H.; Roth, A.; Martínez-Romero, E. Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J. Evol. Biol. 2012, 25, 2357–2368. [Google Scholar] [CrossRef]
- Michalik, K.; Szklarzewicz, T.; Kalandyk-Kołodziejczyk, M.; Jankowska, W.; Michalik, A. Bacteria belonging to the genus Burkholderia are obligatory symbionts of the eriococcids Acanthococcus aceris Signoret, 1875 and Gossyparia spuria (Modeer, 1778) (Insecta, Hemiptera, Coccoidea). Arthropod. Struct. Dev. 2016, 45, 265–272. [Google Scholar] [CrossRef]
- Chong, R.A.; Moran, N.A. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 2018, 12, 898–908. [Google Scholar] [CrossRef] [Green Version]
- Wernegreen, J.J. Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry. Exp. Cell Res. 2017, 358, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Fukatsu, T.; Ishikawa, H. Phylogenetic position of yeast-like symbiont of Hamiltonaphis styraci (Homoptera, Aphididae) based on 18S rDNA sequence. Insect Biochem. Mol. Biol. 1996, 4, 383–388. [Google Scholar] [CrossRef]
- Gruwell, M.E.; Hardy, N.B.; Gullan, P.J.; Dittmar, K. Evolutionary relationships among primary endosymbionts of the mealybug subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl. Environ. Microbiol. 2010, 76, 7521–7525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, N.A.; Dunbar, H.E. Sexual acquisition of beneficial symbionts in aphids. Proc. Natl. Acad. Sci. USA 2006, 103, 12803–12806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, N.A.; Yun, Y. Experimental replacement of an obligate insect symbiont. Proc. Natl. Acad. Sci. USA 2015, 112, 2093–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernegreen, J.J. In it for the long haul: Evolutionary consequences of persistent endosymbiosis. Curr. Opin. Genet. Dev. 2017, 47, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Leonard, S.P.; Li, Y.; Moran, N.A. Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proc. Natl. Acad. Sci. USA 2019, 116, 24712–24718. [Google Scholar] [CrossRef]
- Wernegreen, J.J. Mutualism meltdown in insects: Bacteria constrain thermal adaptation. Curr. Opin. Microbiol. 2012, 15, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Corbin, C.; Heyworth, E.R.; Ferrari, J.; Hurst, G.D.D. Heritable symbionts in a world of varying temperature. Heredity 2017, 118, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Ratzka, C.; Gross, R.; Feldhaar, H. Endosymbiont Tolerance and Control within Insect Hosts. Insects 2012, 3, 553–572. [Google Scholar] [CrossRef] [Green Version]
- Laughton, A.M.; Garcia, J.R.; Gerardo, N.M. Condition-dependent alteration of cellular immunity by secondary symbionts in the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 2016, 86, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, D.B.; Nakabachi, A.; Richards, S.; Qu, J.; Murali, S.C.; Gibbs, R.A.; Moran, N.A. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 2014, 31, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, M.E.; Berg, O.G. Muller’s ratchet in symbiont populations. Genetica 2007, 130, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Ok, S.; Smith, S.; Snoeck, K.; Day, J.P.; Jiggins, F.M. Should symbionts be nice or selfish? antiviral effects of Wolbachia are costly but reproductive parasitism is not. PLoS Pathog. 2015, 11, e1005021. [Google Scholar] [CrossRef] [PubMed]
- Uchi, N.; Fukudome, M.; Nozaki, N.; Suzuki, M.; Osuki, K.-I.; Shigenobu, S.; Uchiumi, T. Antimicrobial Activities of Cysteine-rich Peptides Specific to Bacteriocytes of the Pea Aphid Acyrthosiphon pisum. Microbes Environ. 2019, 34, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Ankrah, N.Y.D.; Luan, J.; Douglas, A.E. Cooperative Metabolism in a Three-Partner Insect-Bacterial Symbiosis Revealed by Metabolic Modeling. J. Bacteriol. 2017, 199, e00872-16. [Google Scholar] [CrossRef] [Green Version]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef]
- Naranjo, S.E.; Gibson, R.L. Phytophagy in predaceous Heteroptera: Effects on life history and population dynamics. In Zoophytophagous Heteroptera: Implications for Life History and Integrated Pest Management; Alomar, O., Wiedenmann, R.N., Eds.; Entomological Society of America: Lanham, MD, USA, 1996; pp. 57–93. [Google Scholar]
- Weirauch, C.; Schuh, R.T.; Cassis, G.; Wheeler, W.C. Revisiting habitat and lifestyle transitions in Heteroptera (Insecta: Hemiptera): Insights from a combined morphological and molecular phylogeny. Cladistics 2018, 35, 67–105. [Google Scholar] [CrossRef] [Green Version]
- Cobben, R.H. On the original feeding habits of the Hemiptera (insecta): A reply to merrill sweet. Ann. Entomol. Soc. Am. 1979, 72, 711–715. [Google Scholar] [CrossRef]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef] [PubMed]
- Panizzi, A.R.; Grazia, J. (Eds.) True Bugs (Heteroptera) of the Neotropics. Entomology in Focus, 1st ed.; Springer: Dordrecht, The Netherlands, 2015; Volume 2. [Google Scholar]
- Kuechler, S.M.; Renz, P.; Dettner, K.; Kehl, S. Diversity of symbiotic organs and bacterial endosymbionts of lygaeoid bugs of the families blissidae and lygaeidae (Hemiptera: Heteroptera: Lygaeoidea). Appl. Environ. Microbiol. 2012, 78, 2648–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, H.; Kawasaki, K.; Takeishi, K.; Noda, H. Symbiont of the stink bug Plautia stali synthesizes rough-type lipopolysaccharide. Microbiol Res. 2011, 167, 48–54. [Google Scholar] [CrossRef]
- Itoh, H.; Jang, S.; Takeshita, K.; Ohbayashi, T.; Ohnishi, N.; Meng, X.-Y.; Mitani, Y.; Kikuchi, Y. Host-symbiont specificity determined by microbe-microbe competition in an insect gut. Proc. Natl. Acad. Sci. USA 2019, 116, 22673–22682. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tada, A.; Kikuchi, Y.; Hosokawa, T.; Musolin, D.L.; Fujisaki, K.; Fukatsu, T. Obligate association with gut bacterial symbiont in Japanese populations of the southern green stinkbug Nezara viridula (Heteroptera: Pentatomidae). Appl. Entomol. Zool. 2011, 46, 483–488. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hosokawa, T.; Nikoh, N.; Fukatsu, T. Gut symbiotic bacteria in the cabbage bugs Eurydema rugosa and Eurydema dominulus (Heteroptera: Pentatomidae). Appl. Entomol. Zool. 2012, 47, 1–8. [Google Scholar] [CrossRef]
- Salem, H.; Kreutzer, E.; Sudakaran, S.; Kaltenpoth, M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 2013, 15, 1956–1968. [Google Scholar] [CrossRef]
- Boucias, D.G.; Garcia-Maruniak, A.; Cherry, R.; Lu, H.; Maruniak, J.E.; Lietze, V.-U. Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis. FEMS Microbiol. Ecol. 2012, 82, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Bistolas, K.S.I.; Sakamoto, R.I.; Fernandes, J.A.M.; Goffredi, S.K. Symbiont polyphyly, co-evolution, and necessity in pentatomid stinkbugs from Costa Rica. Front. Microbiol. 2014, 5, 349. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, T.; Ishii, Y.; Nikoh, N.; Fujie, M.; Satoh, N.; Fukatsu, T. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 2016, 1, 15011. [Google Scholar] [CrossRef] [PubMed]
- Kaiwa, N.; Hosokawa, T.; Nikoh, N.; Tanahashi, M.; Moriyama, M.; Meng, X.-Y.; Maeda, T.; Yamaguchi, K.; Shigenobu, S.; Ito, M.; et al. Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr. Biol. 2014, 24, 2465–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikoh, N.; Hosokawa, T.; Oshima, K.; Hattori, M.; Fukatsu, T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol. Evol. 2011, 3, 702–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küchler, S.M.; Dettner, K.; Kehl, S. Molecular characterization and localization of the obligate endosymbiotic bacterium in the birch catkin bug Kleidocerys resedae (Heteroptera: Lygaeidae, Ischnorhynchinae). FEMS Microbiol. Ecol. 2010, 73, 408–418. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, Y.; Kikuchi, Y.; Hosokawa, T.; Koga, R.; Meng, X.-Y.; Kamagata, Y.; Nikoh, N.; Fukatsu, T. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. 2012, 6, 397–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, T.J. Phylogenetic Analysis of Family Groups within the Infraorder Pentatomomorpha (Hemiptera: Heteroptera), with Emphasis on the Lygaeoidea. Ann. Entomol. Soc. Am. 1997, 90, 275–301. [Google Scholar] [CrossRef]
- Kuechler, S.M.; Matsuura, Y.; Dettner, K.; Kikuchi, Y. Phylogenetically Diverse Burkholderia Associated with Midgut Crypts of Spurge Bugs, Dicranocephalus spp. (Heteroptera: Stenocephalidae). Microbes Environ. 2016, 31, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Foottit, R.G.; Adler, P.H. Insect Biodiversity: Science and Society; Foottit, R.G., Adler, P.H., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2017. [Google Scholar]
- Sudakaran, S.; Retz, F.; Kikuchi, Y.; Kost, C.; Kaltenpoth, M. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 2015, 9, 2587–2604. [Google Scholar] [CrossRef] [Green Version]
- Colman, D.R.; Toolson, E.C.; Takacs-Vesbach, C.D. Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 2012, 21, 5124–5137. [Google Scholar] [CrossRef]
- Hypsa, V.; Aksoy, S. Phylogenetic characterization of two transovarially transmitted endosymbionts of the bedbug Cimex lectularius (Heteroptera: Cimicidae). Insect Mol. Biol. 1997, 6, 301–304. [Google Scholar] [CrossRef]
- Husseneder, C.; Park, J.-S.; Howells, A.; Tikhe, C.V.; Davis, J.A. Bacteria Associated with Piezodorus guildinii (Hemiptera: Pentatomidae), With Special Reference to Those Transmitted by Feeding. Environ Entomol. 2017, 46, 159–166. [Google Scholar] [PubMed]
- Onchuru, T.O.; Martinez, A.J.; Kaltenpoth, M. The cotton stainer’s gut microbiota suppresses infection of a cotransmitted trypanosomatid parasite. Mol. Ecol. 2018, 27, 3408–3419. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Salem, H.; Marz, M.; Vogel, H.; Kaltenpoth, M. Transcriptomic immune response of the cotton stainer Dysdercus fasciatus to experimental elimination of vitamin-supplementing intestinal symbionts. PLoS ONE 2014, 9, e114865. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 10257–10262. [Google Scholar] [CrossRef] [Green Version]
- Martinez, A.J.; Onchuru, T.O.; Ingham, C.S.; Sandoval-Calderón, M.; Salem, H.; Deckert, J.; Kaltenpoth, M. Angiosperm to Gymnosperm host-plant switch entails shifts in microbiota of the Welwitschia bug, Probergrothius angolensis (Distant, 1902). Mol. Ecol. 2019, 28, 5172–5187. [Google Scholar] [CrossRef]
- Schuh, R.T.; Slater, J.A. True Bugs of the World (Hemiptera:Heteroptera): Classification and Natural History; Cornell University Press: Ithaca, NY, USA, 1995. [Google Scholar]
- Hwang, W.S.; Weirauch, C. Evolutionary history of assassin bugs (insecta: Hemiptera: Reduviidae): Insights from divergence dating and ancestral state reconstruction. PLoS ONE 2012, 7, e45523. [Google Scholar] [CrossRef] [Green Version]
- Capinera, J.L.E. Encyclopedia of Entomology, 1st ed.; Springer: Dordrecht, The Netherlands, 2006; Volume 3, p. 2580. [Google Scholar]
- Duncan, J.T. On a Bactericidal Principle present in the Alimentary Canal of Insects and Arachnids. Parasitology 1926, 18, 238–252. [Google Scholar] [CrossRef]
- Vallejo, G.A.; Guhl, F.; Schaub, G.A. Triatominae-Trypanosoma cruzi/T. rangeli: Vector-parasite interactions. Acta Tropica 2009, 110, 137–147. [Google Scholar] [CrossRef]
- Durvasula, R.V.; Sundaram, R.K.; Kirsch, P.; Hurwitz, I.; Crawford, C.V.; Dotson, E.; Beard, C.B. Genetic transformation of a Corynebacterial symbiont from the Chagas disease vector Triatoma infestans. Exp. Parasitol. 2008, 119, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Eichler, S.; Reintjes, N.; Jung, M.; Yassin, A.F.; Schaal, K.P. Identification of bacterial isolates and symbionts from wild populations of Triatoma infestans and T. sordida. Memórias Instituto Oswaldo Cruz 1996, 91, 25. [Google Scholar]
- Yassin, A.F. Rhodococcus triatomae sp. nov., isolated from a blood-sucking bug. Int. J. Syst. Evol. Microbiol. 2005, 55, 1575–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodchild, A.J. The bacteria associated with Triatoma infestans and some other species of Reduviidae. Parasitology 1955, 45, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Erikson, D. The Pathogenic Aerobic Organisms of the Actinomyces Group. MRC 1935, 203, 5–32. [Google Scholar]
- Lake, P.; Friend, W.G. The use of artificial diets to determine some of the effects of Nocardia rhodnii on the development of Rhodnius prolixus. J. Insect Physiol. 1968, 14, 543–562. [Google Scholar] [CrossRef]
- Harington, J.S. Studies on Rhodnius prolixus: Growth and development of normal and sterile bugs, and the symbiotic relationship. Parasitology 1960, 50, 279–286. [Google Scholar] [CrossRef]
- Gumpert, J. Untersuchungen über die Symbiose von Tieren mit Pilzen und Bakterien X. Die Symbiose der Triatominen 2. Infektion symbiontenfreier Triatominen mit symbiontischen und saprophytischen Mikroorganismen und gemeinsame Eigenschaften der symbiontischen Stämme. Zeitschrift Allgemeine Mikrobiologie 1962, 2, 290–302. [Google Scholar] [CrossRef]
- Nyirady, S.A. The germfree culture of three species of Triatominae: Triatoma protracta (Uhler), Triatoma rubida (Uhler) and Rhodnius prolixus Stål. J. Med. Entomol. 1973, 10, 417–448. [Google Scholar] [CrossRef]
- Bewig, F.; Schwartz, W. Untersuchungen über die symbiose von tieren mit pilzen und bakterien. Archiv. Mikrobiol. 1956, 24, 174–208. [Google Scholar] [CrossRef]
- Nyirady, S.A. The Intestinal Flora of Triatoma protracta protracta (Uhler)(Hemiptera: Reduviidae) Including New Bacterial Isolation Techniques. Ph.D. Thesis, Loma Linda University, Loma Linda, CA, USA, 1969. [Google Scholar]
- Gumpert, J. Die Funktion der symbiontischen Bakterien in den Triatominen. Zentralblatt Bakterilogie 1962, 315–318. [Google Scholar]
- Cavanagh, P.; Marsden, P.D. Bacteria isolated from the gut of some Reduviid bugs. Trans. R. Soc. Trop. Med. Hyg. 1969, 63, 415–416. [Google Scholar] [CrossRef]
- Weurman, C. Investigations concerning the symbiosis of bacteria in Triatoma infestans (KLUG). Antonie Van Leeuwenhoek 1946, 11, 129–138. [Google Scholar] [CrossRef]
- Harington, J.S. Synthesis of Thiamine and Folic Acid by Nocardia rhodnii, the Micro-symbiont of Rhodnius prolixus. Nature 1960, 188, 1027–1028. [Google Scholar] [CrossRef] [PubMed]
- Gumpert, J.; Schwartz, W. Untersuchungen über die Symbiose von Tieren mit Pilzen und Bakterien X. Die Symbiose der Triatominen 3. Pantothensäurelieferung als Funktion der Symbionten. Zeitschrift Allgemeine Mikrobiologie 1963, 3, 1–14. [Google Scholar] [CrossRef]
- Friend, W.G.; Cartwright, E. A Practical Apparatus for Feeding Artificial Diets to All Stages of Rhodnius prolixus Stål. Can. Entomol. 1963, 95, 362–364. [Google Scholar] [CrossRef]
- Pachebat, J.A.; van Keulen, G.; Whitten, M.M.A.; Girdwood, S.; Del Sol, R.; Dyson, P.J.; Facey, P.D. Draft Genome Sequence of Rhodococcus rhodnii Strain LMG5362, a Symbiont of Rhodnius prolixus (Hemiptera, Reduviidae, Triatominae), the Principle Vector of Trypanosoma cruzi. Genome Announc. 2013, 1, e00329-13. [Google Scholar] [CrossRef] [Green Version]
- Hill, P.; Campbell, J.A.; Petrie, I.A. Rhodnius prolixus and its symbiotic actinomycete: A microbiological, physiological and behavioural study. Proc. R. Soc. Lond. B. Biol. Sci. 1976, 194, 501–525. [Google Scholar]
- Azambuja, P.; Feder, D.; Garcia, E.S. Isolation of Serratia marcescens in the midgut of Rhodnius prolixus: Impact on the establishment of the parasite Trypanosoma cruzi in the vector. Exp. Parasitol. 2004, 107, 89–96. [Google Scholar] [CrossRef]
- Auden, D.T. Studies on the development of Rhodnius prolixus and the effects of its symbiote Nocardia rhodnii. J. Med. Entomol. 1974, 11, 68–71. [Google Scholar] [CrossRef]
- Gomes, J.E.; Azambuja, P.; Garcia, E.S. Comparative studies on the growth and reproductive performances of Rhodnius prolixus reared on different blood sources. Memórias Instituto Oswaldo Cruz 1990, 85, 299–304. [Google Scholar] [CrossRef] [Green Version]
- Guarneri, A.A.; Pereira, M.H.; Diotaiuti, L. Influence of the blood meal source on the development of Triatoma infestans, Triatoma brasiliensis, Triatoma sordida, and Triatoma pseudomaculata (Heteroptera, Reduviidae). J. Med. Entomol. 2000, 37, 373–379. [Google Scholar] [CrossRef]
- Diotaiuti, L.; Dias, J.C. Estudocomparativo do ciclo evolutivo de Rhodnius neglectus alimentados em pombos oucamundongos. Rev. Soc. Bras. Med. Trop. 1987, 20, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Braga, M.V.; Prata, K.C.; Brazil, R.P. Biology of nymphs of Rhodnius robustus Larrousse, 1927 (Hemiptera, Reduviidae), fed on pigeon or on Swiss mouse blood in laboratory conditions. Rev. Bras. Biol. 1999, 59, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Nattero, J.; Rodríguez, C.S.; Crocco, L. Effects of blood meal source on food resource use and reproduction in Triatoma patagonica Del Ponte (Hemiptera, Reduviidae). J. Vector Ecol. 2013, 38, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Emmanuelle-Machado, P.; Koerich, L.B.; Joukoski, D.D.B.; Carvalho-Pinto, C.J.; Grisard, E.C.; Steindel, M. Biology of Triatoma klugi Carcavallo, Jurberg, Lent & Galvão 2001 (Heteroptera: Reduviidae) under laboratory conditions: Effects of distinct blood sources and susceptibility to Trypanosoma cruzi and Trypanosoma rangeli. Memorias Instituto Oswaldo Cruz 2002, 97, 583–587. [Google Scholar]
- Martínez-Ibarra, J.A.; Novelo López, M.; del Rosario Hernández Robles, M.; Grant Guillén, Y. Influence of the blood meal source on the biology of Meccus picturatus Usinger 1939 (Hemiptera: Reduviidae: Triatominae) under laboratory conditions. Memorias Instituto Oswaldo Cruz 2003, 98, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Ibarra, J.A.; Alejandre-Aguilar, R.; Torres-Morales, A.; Trujillo-García, J.C.; Nogueda-Torres, B.; Trujillo-Contreras, F. Biology of three species of the Meccus phyllosomus complex (Hemiptera: Reduviidae: Triatominae) fed on blood of hens and rabbits. Memorias Instituto Oswaldo Cruz 2006, 101, 787–794. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Ibarra, J.A.; Nogueda-Torres, B.; Meraz-Medina, T.; Diaz-Chavez, R.; Virgen-Cobian, C.J.; Quirarte-Brambila, M. Advantageous Feeding on Different Blood Meal Sources by the Chagas Disease Vector Triatoma barberi (Hemiptera: Reduviidae). J. Med. Entomol. 2019, 56, 1565–1570. [Google Scholar] [CrossRef]
- Lehane, M.J. Managing the blood meal. In The Biology of Blood-Sucking in Insects; Cambridge University Press: Cambridge, UK, 2005; pp. 84–115. [Google Scholar]
- Bing, X.; Attardo, G.M.; Vigneron, A.; Aksoy, E.; Scolari, F.; Malacrida, A.; Weiss, B.L.; Aksoy, S. Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: Transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proc. Biol. Sci. 2017, 284, 20170360. [Google Scholar] [CrossRef] [Green Version]
- Santos-Garcia, D.; Juravel, K.; Freilich, S.; Zchori-Fein, E.; Latorre, A.; Moya, A.; Morin, S.; Silva, F.J. To B or not to B: Comparative genomics suggests Arsenophonus as a source of B vitamins in whiteflies. Front. Microbiol. 2018, 9, 2254. [Google Scholar] [CrossRef]
- Sassera, D.; Epis, S.; Pajoro, M.; Bandi, C. Microbial symbiosis and the control of vector-borne pathogens in tsetse flies, human lice, and triatomine bugs. Pathog. Glob. Health 2013, 107, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Yosef, M.; Rot, A.; Mahagna, M.; Kapri, E.; Behar, A.; Gottlieb, Y. Coxiella-Like Endosymbiont of Rhipicephalus sanguineus Is Required for Physiological Processes During Ontogeny. Front. Microbiol. 2020, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Boyd, B.M.; Reed, D.L. Taxonomy of lice and their endosymbiotic bacteria in the post-genomic era. Clin. Microbiol. Infect. 2012, 18, 324–331. [Google Scholar] [CrossRef] [Green Version]
- Boyd, B.M.; Allen, J.M.; Nguyen, N.-P.; Vachaspati, P.; Quicksall, Z.S.; Warnow, T.; Mugisha, L.; Johnson, K.P.; Reed, D.L. Primates, lice and bacteria: Speciation and genome evolution in the symbionts of hominid lice. Mol. Biol. Evol. 2017, 34, 1743–1757. [Google Scholar] [CrossRef] [PubMed]
- Guizzo, M.G.; Parizi, L.F.; Nunes, R.D.; Schama, R.; Albano, R.M.; Tirloni, L.; Oldiges, D.P.; Vieira, R.P.; Oliveira, W.H.C.; Leite, M.; et al. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci. Rep. 2017, 7, 17554. [Google Scholar] [CrossRef]
- Li, L.-H.; Zhang, Y.; Zhu, D. Effects of antibiotic treatment on the fecundity of Rhipicephalus haemaphysaloides ticks. Parasit. Vectors 2018, 11, 242. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.-Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [Green Version]
- Amanzougaghene, N.; Fenollar, F.; Raoult, D.; Mediannikov, O. Where are we with human lice? A review of the current state of knowledge. Front. Cell Infect. Microbiol. 2019, 9, 474. [Google Scholar] [CrossRef] [Green Version]
- Rio, R.V.M.; Attardo, G.M.; Weiss, B.L. Grandeur alliances: Symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 2016, 32, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, M.; Nikoh, N.; Hosokawa, T.; Fukatsu, T. Riboflavin provisioning underlies wolbachia’s fitness contribution to its insect host. MBio 2015, 6, e01732-15. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. The molecular basis of bacterial-insect symbiosis. J. Mol. Biol. 2014, 426, 3830–3837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Weiss, B.L.; Aksoy, S. Tsetse fly microbiota: Form and function. Front. Cell Infect. Microbiol. 2013, 3, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rio, R.V.M.; Symula, R.E.; Wang, J.; Lohs, C.; Wu, Y.; Snyder, A.K.; Bjornson, R.D.; Oshima, K.; Biehl, B.S.; Perna, N.T.; et al. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. MBio 2012, 3, e00240-11. [Google Scholar] [CrossRef] [Green Version]
- Akman, L.; Yamashita, A.; Watanabe, H.; Oshima, K.; Shiba, T.; Hattori, M.; Aksoy, S. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 2002, 32, 402–407. [Google Scholar] [CrossRef]
- Depeint, F.; Bruce, W.R.; Shangari, N.; Mehta, R.; O’Brien, P.J. Mitochondrial function and toxicity: Role of B vitamins on the one-carbon transfer pathways. Chem. Biol. Interact. 2006, 163, 113–132. [Google Scholar] [CrossRef]
- Schaub, G.A. The effects of trypanosomatids on insects. Adv. Parasitol. 1992, 31, 255–319. [Google Scholar] [PubMed]
- Jensen, C.; Schaub, G.A. Development of Blastocrithidia triatomae (Trypanosomatidae) in Triatoma infestans after vitamin B-supplementation of the blood-diet of the bug. Eur. J. Protistol. 1991, 27, 17–20. [Google Scholar] [CrossRef]
- Watkins, R.P. Host-parasite interaction between Trypanosoma species and Rhodnius prolixus Stal (Hemiptera, Reduviidae). Ph.D. Thesis, University of California, Berkeley, CA, USA, 1969. [Google Scholar]
- Eichler, S.; Schaub, G.A. Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 2002, 100, 17–27. [Google Scholar] [CrossRef]
- Taracena, M.L.; Oliveira, P.L.; Almendares, O.; Umaña, C.; Lowenberger, C.; Dotson, E.M.; Paiva-Silva, G.O.; Pennington, P.M. Genetically modifying the insect gut microbiota to control Chagas disease vectors through systemic RNAi. PLoS Negl. Trop. Dis. 2015, 9, e0003358. [Google Scholar] [CrossRef] [Green Version]
- Mao, M.; Yang, X.; Poff, K.; Bennett, G. Comparative Genomics of the Dual-Obligate Symbionts from the Treehopper, Entylia carinata (Hemiptera: Membracidae), Provide Insight into the Origins and Evolution of an Ancient Symbiosis. Genome Biol. Evol. 2017, 9, 1803–1815. [Google Scholar] [CrossRef] [Green Version]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredricks, D.N. Evidence in microbiome science: Standards for the field (and Lab.). Clin. Infect. Dis. 2020, 1–5. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2004, 70, 4800–4806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; García-González, R.; Schanbacher, F.L.; Morrison, M. Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by Archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2008, 74, 889–893. [Google Scholar] [CrossRef] [Green Version]
- Hypsa, V.; Dale, C. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 1997, 47, 1140–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieran, T.J.; Arnold, K.M.H.; Thomas, J.C.; Varian, C.P.; Saldaña, A.; Calzada, J.E.; Glenn, T.C.; Gottdenker, N.L. Regional biogeography of microbiota composition in the Chagas disease vector Rhodnius pallescens. Parasit. Vectors 2019, 12, 504. [Google Scholar] [CrossRef] [Green Version]
- Waltmann, A.; Willcox, A.C.; Balasubramanian, S.; Borrini Mayori, K.; Mendoza Guerrero, S.; Salazar Sanchez, R.S.; Roach, J.; Condori Pino, C.; Gilman, R.H.; Bern, C.; et al. Hindgut microbiota in laboratory-reared and wild Triatoma infestans. PLoS Negl. Trop. Dis. 2019, 13, e0007383. [Google Scholar] [CrossRef] [Green Version]
- Dumonteil, E.; Ramirez-Sierra, M.-J.; Pérez-Carrillo, S.; Teh-Poot, C.; Herrera, C.; Gourbière, S.; Waleckx, E. Detailed ecological associations of triatomines revealed by metabarcoding and next-generation sequencing: Implications for triatomine behavior and Trypanosoma cruzi transmission cycles. Sci. Rep. 2018, 8, 4140. [Google Scholar] [CrossRef]
- Oliveira, J.L.; Cury, J.C.; Gurgel-Gonçalves, R.; Bahia, A.C.; Monteiro, F.A. Field-collected Triatoma sordida from central Brazil display high microbiota diversity that varies with regard to developmental stage and intestinal segmentation. PLoS Negl. Trop. Dis. 2018, 12, e0006709. [Google Scholar] [CrossRef]
- Orantes, L.C.; Monroy, C.; Dorn, P.L.; Stevens, L.; Rizzo, D.M.; Morrissey, L.; Hanley, J.P.; Rodas, A.G.; Richards, B.; Wallin, K.F.; et al. Uncovering vector, parasite, blood meal and microbiome patterns from mixed-DNA specimens of the Chagas disease vector Triatoma dimidiata. PLoS Negl. Trop. Dis. 2018, 12, e0006730. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.S.; Laport, M.S.; Lorosa, E.S.; Jurberg, J.; Dos Santos, K.R.N.; da Silva Neto, M.A.C.; Rachid, C.T.C.d.C.; Atella, G.C. Bacterial community composition in the salivary glands of triatomines (Hemiptera: Reduviidae). PLoS Negl. Trop. Dis. 2018, 12, e0006739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carels, N.; Gumiel, M.; da Mota, F.F.; de Carvalho Moreira, C.J.; Azambuja, P. A metagenomic analysis of bacterial microbiota in the digestive tract of triatomines. Bioinform. Biol. Insights. 2017, 11, 1177932217733422. [Google Scholar] [CrossRef]
- Montoya-Porras, L.M.; Omar, T.-C.; Alzate, J.F.; Moreno-Herrera, C.X.; Cadavid-Restrepo, G.E. 16S rRNA gene amplicon sequencing reveals dominance of Actinobacteria in Rhodnius pallescens compared to Triatoma maculata midgut microbiota in natural populations of vector insects from Colombia. Acta Tropica 2018, 178, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Villavicencio, B.; Correia, N.; Costa, J.; Haag, K.L. Triatomine bugs, their microbiota and Trypanosoma cruzi: Asymmetric responses of bacteria to an infected blood meal. Parasit. Vectors 2016, 9, 636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumiel, M.; da Mota, F.F.; de Sousa Rizzo, V.; Sarquis, O.; de Castro, D.P.; Lima, M.M.; de Souza Garcia, E.; Carels, N.; Azambuja, P. Characterization of the microbiota in the guts of Triatoma brasiliensis and Triatoma pseudomaculata infected by Trypanosoma cruzi in natural conditions using culture independent methods. Parasit. Vectors 2015, 8, 245. [Google Scholar] [CrossRef]
- Sorfová, P.; Skeríková, A.; Hypsa, V. An effect of 16S rRNA intercistronic variability on coevolutionary analysis in symbiotic bacteria: Molecular phylogeny of Arsenophonus triatominarum. Syst. Appl. Microbiol. 2008, 31, 88–100. [Google Scholar] [CrossRef]
- Mann, A.E.; Mitchell, E.A.; Zhang, Y.; Curtis-Robles, R.; Thapa, S.; Hamer, S.A.; Allen, M.S. Comparison of the Bacterial Gut Microbiome of North American Triatoma spp. With and Without Trypanosoma cruzi. Front. Microbiol. 2020, 11, 364. [Google Scholar] [CrossRef] [Green Version]
- Dumonteil, E.; Pronovost, H.; Bierman, E.F.; Sanford, A.; Majeau, A.; Moore, R.; Herrera, C. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol. Ecol. 2020. [Google Scholar] [CrossRef]
- Vieira, C.S.; Mattos, D.P.; Waniek, P.J.; Santangelo, J.M.; Figueiredo, M.B.; Gumiel, M.; da Mota, F.F.; Castro, D.P.; Garcia, E.S.; Azambuja, P. Rhodnius prolixus interaction with Trypanosoma rangeli: Modulation of the immune system and microbiota population. Parasit. Vectors 2015, 8, 135. [Google Scholar] [CrossRef] [Green Version]
- Castro, D.P.; Moraes, C.S.; Gonzalez, M.S.; Ratcliffe, N.A.; Azambuja, P.; Garcia, E.S. Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS ONE 2012, 7, e36591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandara, A.C.P.; Oliveira, J.H.M.; Nunes, R.D.; Goncalves, R.L.S.; Dias, F.A.; Hecht, F.; Fernandes, D.C.; Genta, F.A.; Laurindo, F.R.M.; Oliveira, M.F.; et al. Amino acids trigger down-regulation of superoxide via TORC pathway in the midgut of Rhodnius prolixus. Biosci. Rep. 2016, 36, e00321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, C.S.; Waniek, P.J.; Mattos, D.P.; Castro, D.P.; Mello, C.B.; Ratcliffe, N.A.; Garcia, E.S.; Azambuja, P. Humoral responses in Rhodnius prolixus: Bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit. Vectors 2014, 7, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, C.S.; Moreira, O.C.; Batista, K.K.S.; Ratcliffe, N.A.; Castro, D.P.; Azambuja, P. The NF-κB Inhibitor, IMD-0354, Affects Immune Gene Expression, Bacterial Microbiota and Trypanosoma cruzi Infection in Rhodnius prolixus Midgut. Front. Physiol. 2018, 9, 1189. [Google Scholar] [CrossRef] [PubMed]
- Vionette do Amaral, R.J. Caracterização Molecular e Funcional das Vias Imunológicas de Sinalização Celular de Rhodnius prolixus, vetor da Doença de Chagas. Ph.D. Thesis, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brasil, 2014. [Google Scholar]
- Perlowagora-Szumlewicz, A.; Muller, C.A.; Moreira, C.J. Studies in search of a suitable experimental insect model for xenodiagnosis of hosts with Chagas’ disease. 4--The reflection of parasite stock in the responsiveness of different vector species to chronic infection with different Trypanosoma cruzi stocks. Revista Saúde Pública 1990, 24, 165–177. [Google Scholar] [CrossRef]
- Peterson, J.K.; Graham, A.L.; Dobson, A.P.; Chávez, O.T. Rhodnius prolixus Life History Outcomes Differ when Infected with Different Trypanosoma cruzi I Strains. Am. J. Trop. Med. Hyg. 2015, 93, 564–572. [Google Scholar] [CrossRef]
- Brenière, S.F.; Waleckx, E.; Barnabé, C. Over Six Thousand Trypanosoma cruzi Strains Classified into Discrete Typing Units (DTUs): Attempt at an Inventory. PLoS Negl. Trop. Dis. 2016, 10, e0004792. [Google Scholar] [CrossRef]
- Vieira, C.S.; Waniek, P.J.; Castro, D.P.; Mattos, D.P.; Moreira, O.C.; Azambuja, P. Impact of Trypanosoma cruzi on antimicrobial peptide gene expression and activity in the fat body and midgut of Rhodnius prolixus. Parasit. Vectors 2016, 9, 119. [Google Scholar] [CrossRef] [Green Version]
- Castro, D.P.; Seabra, S.H.; Garcia, E.S.; de Souza, W.; Azambuja, P. Trypanosoma cruzi: Ultrastructural studies of adhesion, lysis and biofilm formation by Serratia marcescens. Exp Parasitol 2007, 117, 201–207. [Google Scholar] [CrossRef]
- Azambuja, P.; Ratcliffe, N.A.; Garcia, E.S. Towards an understanding of the interactions of Trypanosoma cruzi and Trypanosoma rangeli within the reduviid insect host Rhodnius prolixus. Anais Academia Brasileira Ciências 2005, 77, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Da Mota, F.F.; Castro, D.P.; Vieira, C.S.; Gumiel, M.; de Albuquerque, J.P.; Carels, N.; Azambuja, P. In vitro Trypanocidal Activity, Genomic Analysis of Isolates, and in vivo Transcription of Type VI Secretion System of Serratia marcescens Belonging to the Microbiota of Rhodnius prolixus Digestive Tract. Front. Microbiol. 2019, 9, 3205. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.S.; Genta, F.A.; de Azambuja, P.; Schaub, G.A. Interactions between intestinal compounds of triatomines and Trypanosoma cruzi. Trends Parasitol. 2010, 26, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Ursic-Bedoya, R.J.; Lowenberger, C.A. Rhodnius prolixus: Identification of immune-related genes up-regulated in response to pathogens and parasites using suppressive subtractive hybridization. Dev. Comp. Immunol. 2007, 31, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Kollien, A.H.; Fechner, S.; Waniek, P.J.; Schaub, G.A. Isolation and characterization of a cDNA encoding for a lysozyme from the gut of the reduviid bug Triatoma infestans. Arch. Insect Biochem. Physiol. 2003, 53, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.; Sun, F.; Tew, I.; Schaub, G.; Soukou, C.; Nappi, A.; Ratcliffe, N. Differential modulation of Rhodnius prolixus nitric oxide activities following challenge with Trypanosoma rangeli, T. cruzi and bacterial cell wall components. Insect Biochem. Mol. Biol. 2007, 37, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Buarque, D.S.; Gomes, C.M.; Araújo, R.N.; Pereira, M.H.; Ferreira, R.C.; Guarneri, A.A.; Tanaka, A.S. A new antimicrobial protein from the anterior midgut of Triatoma infestans mediates Trypanosoma cruzi establishment by controlling the microbiota. Biochimie 2016, 123, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.A.C.; Waniek, P.J.; Stock, P.; Mayer, C.; Jansen, A.M.; Schaub, G.A. Sequence characterization and expression patterns of defensin and lysozyme encoding genes from the gut of the reduviid bug Triatoma brasiliensis. Insect Biochem. Mol. Biol. 2006, 36, 547–560. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Genta, F.A.; Sorgine, M.H.F.; Logullo, R.; Mesquita, R.D.; Paiva-Silva, G.O.; Majerowicz, D.; Medeiros, M.; Koerich, L.; Terra, W.R.; et al. An insight into the transcriptome of the digestive tract of the bloodsucking bug, Rhodnius prolixus. PLoS Negl. Trop. Dis. 2014, 8, e2594. [Google Scholar] [CrossRef] [Green Version]
- Ouali, R.; Valentim de Brito, K.C.; Salmon, D.; Bousbata, S. High-Throughput Identification of the Rhodnius prolixus Midgut Proteome Unravels a Sophisticated Hematophagic Machinery. Proteomes 2020, 8, 16. [Google Scholar] [CrossRef]
- Mesquita, R.D.; Vionette-Amaral, R.J.; Lowenberger, C.; Rivera-Pomar, R.; Monteiro, F.A.; Minx, P.; Spieth, J.; Carvalho, A.B.; Panzera, F.; Lawson, D.; et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. USA 2015, 112, 14936–14941. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salcedo-Porras, N.; Umaña-Diaz, C.; de Oliveira Barbosa Bitencourt, R.; Lowenberger, C. The Role of Bacterial Symbionts in Triatomines: An Evolutionary Perspective. Microorganisms 2020, 8, 1438. https://doi.org/10.3390/microorganisms8091438
Salcedo-Porras N, Umaña-Diaz C, de Oliveira Barbosa Bitencourt R, Lowenberger C. The Role of Bacterial Symbionts in Triatomines: An Evolutionary Perspective. Microorganisms. 2020; 8(9):1438. https://doi.org/10.3390/microorganisms8091438
Chicago/Turabian StyleSalcedo-Porras, Nicolas, Claudia Umaña-Diaz, Ricardo de Oliveira Barbosa Bitencourt, and Carl Lowenberger. 2020. "The Role of Bacterial Symbionts in Triatomines: An Evolutionary Perspective" Microorganisms 8, no. 9: 1438. https://doi.org/10.3390/microorganisms8091438