Lactobacillus paracasei A13 and High-Pressure Homogenization Stress Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Preparation
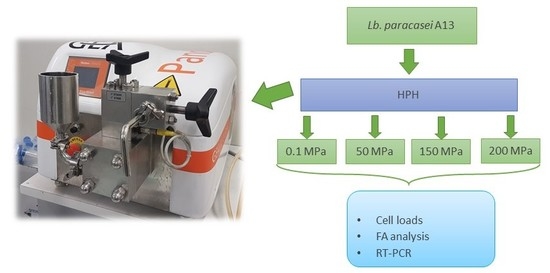
2.2. High-Pressure Homogenization (HPH) Treatment
2.3. Membrane Fatty Acids Analysis
2.4. RNA Extraction and Reverse Transcription
2.5. DNA Extraction
2.6. Genes Selection and Primer Tests on Lactobacillus paracasei A13
2.7. Real-Time PCR (RT-qPCR)
2.8. Relative Gene Expression and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Capra, M.L. Effect of high pressure homogenization on lactic acid bacteria phages and probiotic bacteria phages. Int. Dairy J. 2009, 19, 336–341. [Google Scholar] [CrossRef]
- Dos Santos Aguilar, J.G.; Cristianini, M.; Sato, H.H. Modification of enzymes by use of high-pressure homogenization. Food Res. Int. 2018, 109, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Patrignani, F.; Vannini, L.; Kamdem, S.L.S.; Lanciotti, R.; Guerzoni, M.E. Effect of high pressure homogenization on Saccharomyces cerevisiae inactivation and physico-chemical features in apricot and carrot juices. Int. J. Food Microbiol. 2009, 136, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.; Patrignani, F.; Iucci, L.; Saracino, P.; Guerzoni, M.E. Potential of high pressure homogenization in the control and enhancement of proteolytic and fermentative activities of some Lactobacillus species. Food Chem. 2007, 102, 542–550. [Google Scholar] [CrossRef]
- Show, K.-Y.; Lee, D.-J.; Tay, J.-H.; Lee, T.-M.; Chang, J.-S. Microalgal drying and cell disruption—Recent advances. Bioresour. Technol. 2015, 184, 258–266. [Google Scholar] [CrossRef]
- Betoret, E.; Calabuig-Jiménez, L.; Patrignani, F.; Lanciotti, R.; Dalla Rosa, M. Effect of high pressure processing and trehalose addition on functional properties of mandarin juice enriched with probiotic microorganisms. LWT Food Sci. Technol. 2017, 85, 418–422. [Google Scholar] [CrossRef]
- Patrignani, F.; Lanciotti, R. Applications of high and ultra high pressure homogenization for food safety. Front. Microbiol. 2016, 7, 1132. [Google Scholar] [CrossRef] [Green Version]
- Patrignani, F.; Mannozzi, C.; Tappi, S.; Tylewicz, U.; Pasini, F.; Castellone, V.; Riciputi, Y.; Rocculi, P.; Romani, S.; Caboni, M.F.; et al. (Ultra)-high pressure homogenization potential on the shelf-life and functionality of kiwifruit juice. Front. Microbiol. 2019, 10, 246. [Google Scholar] [CrossRef]
- Zamora, A.; Guamis, B. Opportunities for ultra-high-pressure homogenisation (UHPH) for the food industry. Food Eng. Rev. 2015, 7, 130–142. [Google Scholar] [CrossRef]
- Patrignani, F.; Ndagijimana, M.; Vernocchi, P.; Gianotti, A.; Riponi, C.; Gardini, F.; Lanciotti, R. High-pressure homogenization to modify yeast performance for sparkling wine production according to traditional methods. Am. J. Enol. Vitic. 2013, 64, 258–267. [Google Scholar] [CrossRef]
- Błaszczak, W.; Amarowicz, R.; Górecki, A.R. Antioxidant capacity, phenolic composition and microbial stability of aronia juice subjected to high hydrostatic pressure processing. Innov. Food Sci. Emerg. Technol. 2017, 39, 141–147. [Google Scholar] [CrossRef]
- Diels, A.M.J.; Michiels, C.W. High-Pressure Homogenization as a Non-Thermal Technique for the Inactivation of Microorganisms. Crit. Rev. Microbiol. 2016, 32, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Comuzzo, P.; Calligaris, S.; Iacumin, L.; Ginaldi, F.; Palacios Paz, A.E.; Zironi, R. Potential of high pressure homogenization to induce autolysis of wine yeasts. Food Chem. 2015, 185, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Serrazanetti, D.I.; Patrignani, F.; Russo, A.; Vannini, L.; Siroli, L.; Gardini, F.; Lanciotti, R. Cell membrane fatty acid changes and desaturase expression of Saccharomyces bayanus exposed to high pressure homogenization in relation to the supplementation of exogenous unsaturated fatty acids. Front. Microbiol. 2015, 6, 1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramalla, T.; Aryana, K.J. Some low homogenization pressures improve certain probiotic characteristics of yogurt culture bacteria and Lactobacillus acidophilus LA-K1. J. Dairy Sci. 2011, 94, 3725–3738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabanelli, G.; Patrignani, F.; Vinderola, G.; Reinheimer, J.A.; Gardini, F.; Lanciotti, R. Effect of sub-lethal high pressure homogenization treatments on the in vitro functional and biological properties of lactic acid bacteria. LWT Food Sci. Technol. 2013, 53, 580–586. [Google Scholar] [CrossRef]
- Tabanelli, G.; Patrignani, F.; Gardini, F.; Vinderola, G.; Reinheimer, J.; Grazia, L.; Lanciotti, R. Effect of a sublethal high-pressure homogenization treatment on the fatty acid membrane composition of probiotic lactobacilli. Lett. Appl. Microbiol. 2014, 58, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Burns, P.G.; Patrignani, F.; Tabanelli, G.; Vinderola, G.C.; Siroli, L.; Reinheimer, J.A.; Gardini, F.; Lanciotti, R. Potential of high pressure homogenisation on probiotic Caciotta cheese quality and functionality. J. Funct. Foods 2015, 13, 126–136. [Google Scholar] [CrossRef]
- Tabanelli, G.; Vernocchi, P.; Patrignani, F.; Del Chierico, F.; Putignani, L.; Vinderola, G.; Reinheimer, J.; Gardini, F.; Lanciotti, R. Effects of sub-lethal high-pressure homogenization treatment on the outermost cellular structures and the volatile-molecule profiles of two strains of probiotic lactobacilli. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Suutari, M.; Liukkonen, K.; Laakso, S. Temperature adaptation in yeasts: The role of fatty acids. J. Gen. Microbiol. 1990, 136, 1469–1474. [Google Scholar] [CrossRef] [Green Version]
- Braschi, G.; Serrazanetti, D.I.; Siroli, L.; Patrignani, F.; De Angelis, M.; Lanciotti, R. Gene expression responses of Listeria monocytogenes Scott A exposed to sub-lethal concentrations of natural antimicrobials. Int. J. Food Microbiol. 2018, 286, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, C.; Zúñiga, M. Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology 2012, 158, 1206–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Short technical report processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32, 1372–1379. [Google Scholar] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Tasara, T.; Stephan, R. Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol. Lett. 2007, 269, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Burns, P.; Patrignani, F.; Serrazanetti, D.; Vinderola, G.C.; Reinheimer, J.A.; Lanciotti, R.; Guerzoni, M.E. Probiotic Crescenza Cheese Containing and Lactobacillus acidophilus manufactured with High-Pressure Homogenized milk. J. Dairy Sci. 2008, 91, 500–512. [Google Scholar] [CrossRef] [Green Version]
- Vinderola, G.; Binetti, A.; Burns, P.; Reinheimer, J. Cell Viability and Functionality of Probiotic Bacteria in Dairy Products. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef] [Green Version]
- Tabanelli, G.; Burns, P.; Patrignani, F.; Gardini, F.; Lanciotti, R.; Reinheimer, J.; Vinderola, G. Effect of a non-lethal High Pressure Homogenization treatment on the in vivo response of probiotic lactobacilli. Food Microbiol. 2012, 32, 302–307. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Wang, M.; Du, G.; Chen, J. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. Molecules 2012, 39, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Murínová, S.; Dercová, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 873081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siroli, L.; Braschi, G.; de Jong, A.; Kok, J.; Patrignani, F.; Lanciotti, R. Transcriptomic approach and membrane fatty acid analysis to study the response mechanisms of Escherichia coli to thyme essential oil, carvacrol, 2-(E)-hexanal and citral exposure. J. Appl. Microbiol. 2018, 125, 1308–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Pasqua, R.; Hoskins, N.; Betts, G.; Mauriello, G. Changes in Membrane Fatty Acids Composition of Microbial Cells Induced by Addiction of Thymol, Carvacrol, Limonene, Cinnamaldehyde, and Eugenol in the Growing Media. J. Agric. Food Chem. 2006, 54, 2745–2749. [Google Scholar] [CrossRef]
- Gianotti, A.; Iucci, L.; Guerzoni, M.E.; Lanciotti, R. Effect of acidic conditions on fatty acid composition and membrane fluidity of Escherichia coli strains isolated from Crescenza cheese. Ann. Microbiol. 2009, 59, 603. [Google Scholar] [CrossRef]
- Siroli, L.; Patrignani, F.; Gardini, F.; Lanciotti, R. Effects of sub-lethal concentrations of thyme and oregano essential oils, carvacrol, thymol, citral and trans-2-hexenal on membrane fatty acid composition and volatile molecule profile of Listeria monocytogenes, Escherichia coli and Salmonella enteritidis. Food Chem. 2015, 182, 185–192. [Google Scholar] [CrossRef]
- Yoon, Y.; Lee, H.; Lee, S.; Kim, S.; Choi, K.-H. Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res. Int. 2015, 72, 25–36. [Google Scholar] [CrossRef]
- Royce, L.A.; Yoon, J.M.; Chen, Y.; Rickenbach, E.; Shanks, J.V.; Jarboe, L.R. Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab. Eng. 2015, 29, 180–188. [Google Scholar] [CrossRef]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef]
- Patrignani, F.; Siroli, L.; Serrazanetti, D.I.; Braschi, G.; Betoret, E.; Reinheimer, J.A.; Lanciotti, R. Microencapsulation of functional strains by high pressure homogenization for a potential use in fermented milk. Food Res. Int. 2017, 97, 250–257. [Google Scholar] [CrossRef]
- Granato, D.; Branco, G.F.; Nazzaro, F.; Cruz, A.G.; Faria, J.A.F. Functional foods and nondairy probiotic food development: Trends, concepts, and products. Compr. Rev. Food Sci. Food Saf. 2010, 9, 292–302. [Google Scholar] [CrossRef]
- Terpou, A.; Papadaki, A.; Lappa, I.K.; Kachrimanidou, V.; Bosnea, L.A.; Kopsahelis, N. Probiotics in food systems: Significance and emerging strategies towards improved viability and delivery of enhanced beneficial value. Nutrients 2019, 11, 1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floury, J.; Desrumaux, A.; Lardières, J. Effect of high-pressure homogenization on droplet size distributions and rheological properties of model oil-in-water emulsions. Innov. Food Sci. Emerg. 2000, 1, 127–134. [Google Scholar] [CrossRef]
- Floury, J.; Bellettre, J.; Legrand, J.; Desrumaux, A. Analysis of a new type of high pressure homogeniser. A study of the flow pattern. Procedia Food Sci. 2004, 59, 843–853. [Google Scholar] [CrossRef]
- Pinho, C.R.G.; Franchi, M.A.; Tribst, A.A.L.; Cristianinia, M. Effect of high pressure homogenization process on Bacillus stearothermophilus and Clostridium sporogenes spores in skim milk. Procedia Food Sci. 2011, 1, 869–873. [Google Scholar] [CrossRef] [Green Version]
- Lanciotti, R.; Sinigaglia, M.; Angelini, P.; Guerzoni, M.E. Effects of homogenization pressure on the survival and growth of some food spoilage and pathogenic micro-organisms. Lett. Appl. Microbiol. 1994, 18, 319–322. [Google Scholar] [CrossRef]
- Lanciotti, R.; Gardini, F.; Sinigaglia, M.; Guerzoni, M.E. Effects of growth conditions on the resistance of some pathogenic and spoilage species to high pressure homogenization. Lett. Appl. Microbiol. 1996, 22, 165–168. [Google Scholar] [CrossRef]
- Guerzoni, M.E.; Lanciotti, R.; Cocconcelli, P.S. Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 2001, 147, 2255–2264. [Google Scholar] [CrossRef] [Green Version]
- Dodd, C.E.R.; Sharman, R.L.; Bloomfield, S.F.; Booth, I.R.; Stewart, G.S.A.B. Inimical processes: Bacterial self-destruction and sub-lethal injury. Trends Food Sci. Technol. 1997, 8, 238–241. [Google Scholar] [CrossRef]
- Oktyabrskii, O.N.; Smirnova, G.V. Redox potential changes in bacterial cultures under stress conditions. Microbiology 2012, 81, 131–142. [Google Scholar] [CrossRef]
- Donsì, F.; Annunziata, M.; Ferrari, G. Microbial inactivation by high pressure homogenization: Effect of the disruption valve geometry. J. Food Eng. 2013, 115, 362–370. [Google Scholar] [CrossRef]
- Guerzoni, M.E.; Vannini, L.; Chaves Lopez, C.; Lanciotti, R.; Suzzi, G.; Gianotti, A. Effect of high pressure homogenization on microbial and chemico-physical characteristics of goat cheeses. J. Dairy Sci. 1999, 82, 851–862. [Google Scholar] [CrossRef]
- Lanciotti, R.; Gardini, F.; Sinigaglia, M.; Guerzoni, M.E. Physiological responses to sublethal hydrostatic pressure in Yarrowia lipolytica. Lett. Appl. Microbiol. 1997, 24, 27–32. [Google Scholar] [CrossRef]
- Guerzoni, M.E.; Ferruzzi, M.; Sinigaglia, M.; Criscuoli, G.C. Increased cellular fatty acid desaturation as a possible key factor in thermotolerance in Saccharomyces cerevisiae. Can. J. Microbiol. 1997, 43, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.T.; Khalawan, S.A.; Curran, B.P.G. Cellular lipid composition influences stress activation of the yeast general stress response element (STRE). Microbiology 2000, 146, 877–884. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [Green Version]
- Montanari, C.; Sado Kamdem, S.L.; Serrazanetti, D.I.; Etoa, F.-X.; Guerzoni, M.E. Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 2010, 27, 493–502. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Serrazanetti, D.I.; Guerzoni, M.E.; Corsetti, A.; Vogel, R. Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol. 2009, 26, 700–711. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Liang, N.Y.; Curtis, J.M.; Gänzle, M.G. Characterization of linoleate 10-Hydratase of Lactobacillus plantarum and novel antifungal metabolites. Front. Microbiol. 2016, 7, 1561. [Google Scholar] [CrossRef] [Green Version]
- Montanari, C.; Sado Kamdem, S.L.; Serrazanetti, D.I.; Vannini, L.; Guerzoni, M.E. Oxylipins generation in Lactobacillus helveticus in relation to unsaturated fatty acid supplementation. J. Appl. Microbiol. 2013, 115, 1388–1401. [Google Scholar] [CrossRef] [PubMed]
- Ndagijimana, M.; Vallicelli, M.; Cocconcelli, P.S.; Cappa, F.; Patrignani, F.; Lanciotti, R.; Guerzoni, M.E. Two 2[5H]-Furanones as Possible Signaling Molecules in Lactobacillus helveticus. Appl. Environ. Microbiol. 2006, 72, 6053–6061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannini, L.; Ndagijimana, M.; Saracino, P.; Vernocchi, P.; Corsetti, A.; Vallicelli, M.; Cappa, F.; Cocconcelli, P.S.; Guerzoni, M.E. New signaling molecules in some gram-positive and gram-negative bacteria. Int. J. Food Microbiol. 2007, 120, 25–33. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Cronan, J.E. β-Ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 2003, 278, 51494–51503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szafranska, A.E.; Hitchman, T.S.; Cox, R.J.; Crosby, J.; Simpson, T.J. Kinetic and mechanistic analysis of the malonyl coa:acp transacylase from streptomyces coelicolor indicates a single catalytically competent serine nucleophile at the active site. Biochemistry 2002, 41, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Ogawa, J.; Penaud, S.; Boudebbouze, S.; Ehrlich, D.; van de Guchte, M.; Maguin, E. Rerouting of pyruvate metabolism during acid adaptation in Lactobacillus bulgaricus. Proteomics 2008, 8, 3154–3163. [Google Scholar] [CrossRef]
- Srisukchayakul, P.; Charalampopoulos, D.; Karatzas, K.A. Study on the effect of citric acid adaptation toward the subsequent survival of Lactobacillus plantarum NCIMB 8826 in low pH fruit juices during refrigerated storage. Food Res. Int. 2018, 111, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.-H.; Heath, R.J.; Rock, C.O. β-Ketoacyl-acyl carrier protein synthase iii (fabh) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 2000, 182, 365–370. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Bi, H.; Ma, J.; Hu, Z.; Zhang, W.; Cronan, J.E.; Wang, H. The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 2013, 4, e00613-13. [Google Scholar] [CrossRef] [Green Version]
- Heath, R.J.; Rock, C.O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 1996, 271, 1833–1836. [Google Scholar] [CrossRef] [Green Version]
- Beld, J.; Lee, D.J.; Burkart, M.D. Fatty acid biosynthesis revisited: Structure elucidation and metabolic engineering. Mol. BioSyst. 2015, 11, 38–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, A.C.; Rock, C.O.; White, S.W. The 1.3-Angstrom-Resolution Crystal Structure of β-Ketoacyl-Acyl Carrier Protein Synthase II from Streptococcus pneumoniae. J. Bacteriol. 2003, 185, 4136–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bokhorst-van de Veen, H.; Abee, T.; Tempelaars, M.; Bron, P.A.; Kleerebezem, M.; Marco, M.L. Short- and long-term adaptation to ethanol stress and its cross-protective consequences in Lactobacillus plantarum. Appl. Environ. Microbiol. 2011, 77, 5247–5256. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Zhang, B.; Guo, H.; Cui, J.; Jiang, L.; Song, S.; Sun, M.; Ren, F. Mechanism analysis of acid tolerance response of Bifidobacterium longum subsp. longum BBMN 68 by gene expression profile using RNA-sequencing. PLoS ONE 2012, 7, e50777. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Primer Sequence (5′ > 3′) | Biological Function | MgCl2 (mM) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|
| accA | F: CCGGCGCATATCCTGGCAAA | acetyl-CoA carboxylase carboxyl transferase subunit alpha | 2 | 62 | This study |
| R: ACCTCATCCCCAAACGCCAA | |||||
| accC | F: ACTTCACCTCGCTGCGACCA | acetyl-CoA carboxylase, biotin carboxylase subunit | 2 | 64 | This study |
| R: TGCCACTTCATCGCCAGCTG | |||||
| accD | F: TTGCCCGCACTGCCATTACG | acetyl-CoA carboxylase carboxyl transferase subunit beta | 3 | 62 | This study |
| R: TCGCCTGACCTGTCCACACA | |||||
| fabD | F: GCATTGCCCGATGAAACCGC | malonyl CoA-acyl carrier protein transacylase | 2 | 62 | This study |
| R: GCACTTGTTGACAGGCAGCC | |||||
| fabH | F: TTTGCTGATGGTGCTGGCGG | 3-oxoacyl-ACP-synthase III | 4 | 64 | This study |
| R: ACCGCCCGCCCATTCATTTT | |||||
| fabG | F: GCGTGCGAGTCAATGCGATC | 3-oxoacyl-ACP-reductase | 2 | 62 | This study |
| R: ACCAGAAAACGGGCCGCATG | |||||
| fabZ | F: CGATGCCGGACTTCAAAGGA | (3R)-hydroxymyristoyl-ACP-dehydratase | 2 | 62 | This study |
| R: ACTAAATCCGCACTGCTGGC | |||||
| fabK | F: TTGCTGACGGTCGTGGTGTG | enoyl-ACP-protein reductase | 3 | 62 | This study |
| R: CGGTCGTGTCAGAAAGGGCA | |||||
| fabF | F: TTTACTGGGTGCGGCTGGTG | 3-oxoacyl-ACP-synthase II | 2 | 62 | This study |
| R: ATGCCCGCCGAAACCAAAGG | |||||
| mch | F: CGGTTTTGCGGGTGATCGGT | acyl-ACP thioesterase | 3 | 62 | This study |
| R: TGGTTCCTCAATTCGCGGCA | |||||
| ileS (RG) | F: ACCATTCCGGCTAACTATGG | Isoleucine tRNA ligase | 2 | 64 | Alcántara et al. [22] |
| R: TCAGGATCTTCGGATTTTCC | |||||
| lepA (RG) | F: CACATTGATCACGGGAAGTC | Elongation factor 4 | 2 | 64 | Alcántara et al. [22] |
| R: GTAATGCCACGTTCACGTTC | |||||
| pyrG (RG) | F: AATTGCGCTTTTCACTGATG | CTP synthase | 2 | 64 | Alcántara et al. [22] |
| R: CGAAATGATCGACCACAATC | |||||
| pcrA (RG) | F: CGGCCAATAATGTGATTCAG | ATP-dependent DNA helicase | 2 | 64 | Alcántara et al. [22] |
| R: TCATCAGTTTCGCTTTGAGC |
| Pressure MPa | log CFU/mL ± SD |
|---|---|
| Pre-Treatment | 9.18 ± 0.11 a |
| 0.1 | 9.20 ± 0.09 a |
| 50 | 9.00 ± 0.15 a |
| 150 | 9.16 ± 0.08 a |
| 200 | 9.20 ± 0.12 a |
| MPa | ||||
|---|---|---|---|---|
| Fatty Acid | 0.1 (Control) | 50 | 150 | 200 |
| C12:0 | 100.0 ± 3.4 a | 120.5 ± 3.2 b | 180.1 ± 5.4 c | 181.7 ± 6.2 c |
| 2-OH C12:0 | 1.8 ± 0.1 | - | - | - |
| C13:0 | 2.9 ± 0.1 a | 11.8 ± 0.4 b | 61.1 ± 2.5 d | 46.5 ± 1.6 c |
| 3-OH C12:0 | 3.4 ± 0.1 a | 3.3 ± 0.2 a | - | - |
| C14:0 | 18.2 ± 0.5 a | 18.5 ± 0.9 a | 38.9 ± 1.9 b | 38.3 ± 2.1 b |
| C15:0 | 10.0 ± 0.2 | - | - | - |
| 2-OH C14:0 | 22.5 ± 1.1 a | 11.0 ± 0.8 b | 22.4 ± 1.8 a | - |
| 3-OH C14:0 | 9.1 ± 0.6 a | 4.2 ± 0.2 b | - | - |
| i-16:0 | 17.3 ± 0.8 a | 11.9 ± 0.9 b | - | 14.0 ± 0.9 b |
| C16:1 9 cis | 30.5 ± 1.8 a | 35.1 ± 1.5 a | 59.3 ± 2.3 b | 57.8 ± 2.2 b |
| C16:0 | 114.6 ± 6.3 a | 101.7 ± 5.2 a | 145.8 ± 5.2 b | 147.0 ± 7.9 b |
| C18:1 9 cis | 150.4 ± 7.5 a | 170.2 ± 8.9 a | 339.4 ± 18.9 c | 280.6 ± 15.3 b |
| C18:1 9 trans | 14.54 ± 1.1 a | 19.3 ± 1.9 b | 33.1 ± 2.5 d | 26.2 ± 1.6 c |
| C18:0 | - | 28.1 ± 4.3 b | 56.5 ± 4.6 c | 34.3 ± 2.3 d |
| C19Cyc9+C19cyc11 | 152.1 ± 6.8 a | 155.3 ± 7.5 a | 292.9 ± 11.5 b | 307.5 ± 18.6 b |
| UFA 1 | 195.4 ± 12.4 a | 224.5 ± 13.3 b | 431.8 ± 22.6 d | 364.6 ± 24.6 c |
| SFA 2 | 451.9 ± 19.8 a | 466.2 ± 15.4 a | 797.6 ± 39.8 b | 769.3 ± 45.7 b |
| UFA/SFA 3 | 0.43 ± 0.2 | 0.48 ± 0.1 | 0.54 ± 0.3 | 0.47 ± 0.3 |
| CL 4 | 16.1 ± 0.1 | 16.4 ± 0.2 | 16.6 ± 0.2 | 16.8 ± 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siroli, L.; Braschi, G.; Rossi, S.; Gottardi, D.; Patrignani, F.; Lanciotti, R. Lactobacillus paracasei A13 and High-Pressure Homogenization Stress Response. Microorganisms 2020, 8, 439. https://doi.org/10.3390/microorganisms8030439
Siroli L, Braschi G, Rossi S, Gottardi D, Patrignani F, Lanciotti R. Lactobacillus paracasei A13 and High-Pressure Homogenization Stress Response. Microorganisms. 2020; 8(3):439. https://doi.org/10.3390/microorganisms8030439
Chicago/Turabian StyleSiroli, Lorenzo, Giacomo Braschi, Samantha Rossi, Davide Gottardi, Francesca Patrignani, and Rosalba Lanciotti. 2020. "Lactobacillus paracasei A13 and High-Pressure Homogenization Stress Response" Microorganisms 8, no. 3: 439. https://doi.org/10.3390/microorganisms8030439