Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth
Abstract
:1. Introduction
2. Results
2.1. Effect of Phycocyanin on the Morphology and Growth of the NSCLC A549 Cell Line
2.2. RNA Sequencing Data Analysis: Quality Control, Assembly, and Mapping
2.3. Functional Annotation and Gene Ontology Classification
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Gene Ontology Analysis of Differentially Expressed Genes
2.6. KEGG Pathway Analysis of Differentially Expressed Genes
2.7. Protein–Protein Interaction (PPI) Network Analysis of Differentially Expressed Genes
2.8. qRT-PCR Validation of Differentially Expressed Genes
2.9. Pathway Detection in A549 Cells after Phycocyanin Treatment
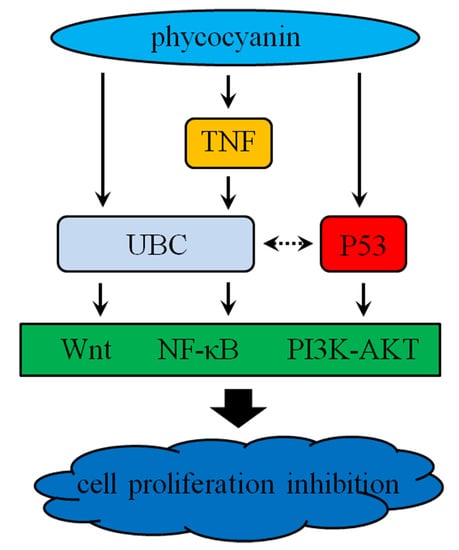
3. Discussion
4. Materials and Methods
4.1. Materials, Cell Line and Culture Conditions
4.2. Cell Survival Rate Assay
4.3. Cell Proliferation Assay
4.4. RNA Extraction and cDNA Library Construction
4.5. Transcriptome Sequencing and Bioinformatics Analysis
4.6. Quantitative RT-PCR (qRT-PCR) on Gene Expressions
4.7. Western Blotting
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, C.; Hu, T.; Wang, W. S-1-based concurrent chemoradiotherapy in the treatment of locally advanced non-small cell lung cancer: A systematic review and meta-analysis protocol. Medicine 2018, 97, e0397. [Google Scholar] [CrossRef] [PubMed]
- Carrizosa, D.R.; Gold, K.A. New strategies in immunotherapy for non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 553–559. [Google Scholar] [PubMed]
- Chen, Y.Y.; Huang, T.W.; Tsai, W.C.; Lin, L.F.; Cheng, J.B.; Chang, H.; Lee, S.C. Risk factors of postoperative recurrences in patients with clinical stage I NSCLC. World J Surg. Oncol. 2014, 12, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Huang, Y.; Zhang, R.; Cai, T.; Cai, Y. Medical application of Spirulina platensis derived C-phycocyanin. Evid. Based Complement. Alternat. Med. 2016, 2016, 7803846. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chu, X.; Gao, M.; Li, W. Apoptotic mechanism of MCF-7 breast cells in vivo and in vitro induced by photodynamic therapy with C-phycocyanin. Acta Biochim. Biophys. Sin. 2010, 42, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Thangam, R.; Suresh, V.; Asenath Princy, W.; Rajkumar, M.; Senthilkumar, N.; Gunasekaran, P.; Rengasamy, R.; Anbazhagan, C.; Kaveri, K.; Kannan, S. C-Phycocyanin from Oscillatoria tenuis exhibited an antioxidant and in vitro antiproliferative activity through induction of apoptosis and G0/G1 cell cycle arrest. Food Chem. 2013, 140, 262–272. [Google Scholar] [CrossRef]
- Nemoto-Kawamura, C.; Hirahashi, T.; Nagai, T.; Yamada, H.; Katoh, T.; Hayashi, O. Phycocyanin enhances secretary IgA antibody response and suppresses allergic IgE antibody response in mice immunized with antigen-entrapped biodegradable microparticles. J. Nutr. Sci. Vitaminol. 2004, 50, 129–136. [Google Scholar] [CrossRef]
- Jensen, G.S.; Attridge, V.L.; Beaman, J.L.; Guthrie, J.; Ehmann, A.; Benson, K.F. Antioxidant and anti-inflammatory properties of an aqueous cyanophyta extract derived from Arthrospira platensis: Contribution to bioactivities by the non-phycocyanin aqueous fraction. J. Med. Food 2015, 18, 535–541. [Google Scholar] [CrossRef]
- Eriksen, N.T. Production of phycocyanin—A pigment with applications in biology, biotechnology, foods and medicine. Appl. Microbiol. Biotechnol. 2008, 80, 1–14. [Google Scholar] [CrossRef]
- Liao, G.Y.; Gao, B.; Gao, Y.N.; Yang, X.G.; Cheng, X.D.; Ou, Y. Phycocyanin inhibits tumorigenic potential of pancreatic cancer cells: Role of apoptosis and autophagy. Sci. Rep. 2016, 6, 34564. [Google Scholar] [CrossRef] [PubMed]
- Saini, M.K.; Sanyal, S.N. Targeting angiogenic pathway for chemoprevention of experimental colon cancer using C-Phycocyanin as cyclooxygenase-2 inhibitor. Biochem. Cell Biol. 2014, 92, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Fu, X.; Huang, W.; Li, C.X.; Wang, X.Y.; Huang, B. Photodynamic effect and mechanism study of selenium-enriched phycocyanin from Spirulina platensis against liver tumours. J. Photochem. Photobiol. 2018, 180, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Minic, S.L.; Stanic-Vucinic, D.; Mihailovic, J.; Krstic, M.; Nikolic, M.R.; Cirkovic Velickovic, T. Digestion by pepsin releases biologically active chromopeptides from C-phycocyanin, a blue-colored biliprotein of microalga Spirulina. J. Proteomics 2016, 147, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, M.H.; Chu, X.M.; Teng, L.; Lv, C.Y.; Yang, P.; Yin, Q.F. The synergistic antitumor effects of all-trans retinoic acid and C-Phycocyanin on the lung cancer A549 cells in vitro and in vivo. Eur. J. Pharmacol. 2015, 749, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, M.H.; Lv, C.Y.; Yang, P.; Yin, Q.F. Study of the synergistic effects of all-transretinoic acid and C-Phycocyanin on the growth and apoptosis of A549 cells. Eur. J. Cancer Prev. 2016, 25, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Baudelet, P.H.; Gagez, A.L.; Berard, J.B.; Juin, C.; Bridiau, N.; Kaas, R.; Thiery, V.; Cadoret, J.P.; Picot, L. Antiproliferative activity of Cyanophora paradoxa pigments in melanoma, breast and lung cancer cells. Mar. Drugs 2013, 11, 4390–4406. [Google Scholar] [CrossRef]
- Bingula, R.; Dupuis, C.; Pichon, C.; Berthon, J.Y.; Filaire, M.; Pigeon, L.; Falaire, E. Study of the effects of betaine and/or C-Phycocyanin on the growth of lung cancer A549 cells in vitro and in vivo. J. Oncol. 2016, 2016, 8162952. [Google Scholar] [CrossRef]
- Hao, S.; Yan, Y.; Li, S.; Zhao, L.; Zhang, C.; Liu, L.Y.; Wang, C.T. The in vitro anti-tumor activity of phycocyanin against non-small cell lung cancer cells. Mar. Drugs 2018, 16, 178. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Wilhelm, B.T.; Marguerat, S.; Goodhead, I.; Bähler, J. Defining transcribed regions using RNA-seq. Nat. Protoc. 2010, 5, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.; Wei, W.; Yin, G.; Hettinghouse, A.; Liu, C.; Shi, Y. RNA-seq analysis of interferon inducible p204-mediated network in anti-tumor immunity. Sci. Rep. 2018, 8, 6495. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, C.M.; Hu, Y.; Daniel, R.; Gaddis, S.; Kittrell, F.; Medina, D. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene 2002, 21, 6366–6376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, J.; Wang, J.; Ji, H.; Lin, C.; Pan, R.; Zhou, L.; Song, Y.; Zhang, E.; Ren, P.; Chen, J.; et al. Transcriptome analysis of phycocyanin inhibitory effects on SKOV-3 cell proliferation. Gene 2016, 585, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Subhashini, J.; Mahipal, S.V.; Reddy, M.C.; Mallikarjuna Reddy, M.; Rachamallu, A.; Reddanna, P. Molecular mechanisms in C-Phycocyanin induced apoptosis in human chronic myeloid leukemia cell line-K562. Biochem. Pharmacol. 2004, 68, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Wiborg, O.; Pedersen, M.S.; Wind, A.; Berglund, L.E.; Marcker, K.A.; Vuust, J. The human ubiquitin multigene family: Some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985, 4, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef]
- Suzuki, S.; Kurata, M.; Abe, S.; Miyazawa, R.; Murayama, T.; Hidaka, M.; Yamamoto, K.; Kitagawa, M. Overexpression of MCM2 in myelodysplastic syndromes: Association with bone marrow cell apoptosis and peripheral cytopenia. Exp. Mol. Pathol. 2012, 92, 160–166. [Google Scholar] [CrossRef]
- Samel, S.A.; Fernandez-Cid, A.; Sun, J.; Riera, A.; Tognetti, S.; Herrera, M.C.; Li, H.; Speck, C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014, 28, 1653–1666. [Google Scholar] [CrossRef]
- Sandrini, G.; Cunsolo, S.; Schuurmans, J.M.; Matthijs, H.C.; Huisman, J. Changes in gene expression, cell physiology and toxicity of the harmful cyanobacterium Microcystis aeruginosa at elevated CO2. Front. Microbiol. 2015, 6, 401. [Google Scholar] [CrossRef]
- Dienst, D.; Georg, J.; Abts, T.; Jakorew, L.; Kuchmina, E.; Borner, T.; Wilde, A.; Duhring, U.; Enke, H.; Hess, W.R. Transcriptomic response to prolonged ethanol production in the cyanobacterium Synechocystis sp. PCC6803. Biotechnol. Biofuels. 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Scie. USA 1980, 77, 1365–1368. [Google Scholar] [CrossRef]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-Guiu, X. NF-kB in development and progression of human cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Roy, N.; Stennicke, H.R.; Van Arsdale, T.; Zhou, Q.; Srinivasula, S.M.; Alnemri, E.S.; Salvesen, G.S.; Reed, J.C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998, 17, 2215–2223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttridge, D.C.; Albanese, C.; Reuther, J.Y.; Pestell, R.G.; Baldwin, A.S., Jr. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999, 19, 5785–5799. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, K.; Verstrepen, L.; Carpentier, I.; Beyaert, R. Linear ubiquitination in NF-kappaB signaling and inflammation: What we do understand and what we do not. Biochem. Pharmacol. 2011, 82, 1057–1065. [Google Scholar] [CrossRef]
- Tai, D.; Wells, K.; Arcaroli, J.; Vanderbilt, C.; Aisner, D.L.; Messersmith, W.A.; Lieu, C.H. Targeting the WNT Signaling Pathway in Cancer Therapeutics. Oncologist 2015, 20, 1189–1198. [Google Scholar] [CrossRef] [Green Version]
- Stewart, D.J. Wnt signaling pathway in non-small cell lung cancer. J. Natl. Cancer Inst. 2014, 106, djt356. [Google Scholar] [CrossRef]
- Han, W.; Lee, H.; Han, J.K. Ubiquitin C-terminal hydrolase37 regulates Tcf7 DNA binding for the activation of Wnt signalling. Sci. Rep. 2017, 7, 42590. [Google Scholar] [CrossRef] [Green Version]
- Tennis, M.A.; Vanscoyk, M.M.; Wilson, L.A.; Kelley, N.; Winn, R.A. Methylation of Wnt7a is modulated by DNMT1 and cigarette smoke condensate in non-small cell lung cancer. PLoS ONE 2012, 7, e32921. [Google Scholar] [CrossRef]
- Winn, R.A.; Marek, L.; Han, S.Y.; Rodriguez, K.; Rodriguez, N.; Hammond, M.; Van Scoyk, M.; Acosta, H.; Mirus, J.; Barry, N.; et al. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J. Biol. Chem. 2005, 280, 19625–19634. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Vousden, K.H. Mechanisms of p53-mediated apoptosis. Cell. Mol. Life Sci. 1999, 55, 28–37. [Google Scholar] [CrossRef]
- Lowe, J.M.; Menendez, D.; Bushel, P.R.; Shatz, M.; Kirk, E.L.; Troester, M.A.; Garantziotis, S.; Fessler, M.B.; Resnick, M.A. p53 and NF-kappaB coregulate proinflammatory gene responses in human macrophages. Cancer Res. 2014, 74, 2182–2192. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, H.S.; Kim, N.G.; Lee, I.; Choi, H.S.; Li, X.Y.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Na, J.M.; et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 2011, 4, ra71. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.G.; O’Neill, E. PI3K/Akt-mediated regulation of p53 in cancer. Biochem. Soc. Trans. 2014, 42, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Marchenko, N.D.; Wolff, S.; Erster, S.; Becker, K.; Moll, U.M. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007, 26, 923–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Krylov, D.M.; Wolf, Y.I.; Rogozin, I.B.; Koonin, E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003, 13, 2229–2235. [Google Scholar] [CrossRef] [PubMed]








| Sample | Raw Reads | Clean Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|---|
| Contorl-1 | 71,748,960 | 66,148,142 | 64,387,922 (97.34%) | 2,099,216 (3.17%) | 62,288,706 (94.17%) |
| Control-2 | 70,062,692 | 64,302,644 | 62,557,790 (97.29%) | 2,054,749 (3.2%) | 60,503,041 (94.09%) |
| Control-3 | 54,566,932 | 52,411,510 | 51,378,836 (98.03%) | 1,624,560 (3.1%) | 49,754,276 (94.93%) |
| PC-1 | 58,518,666 | 56,346,378 | 55,114,355 (97.81%) | 2,257,228 (4.01%) | 52,857,127 (93.81%) |
| PC-2 | 51,947,736 | 49,626,788 | 48,530,748 (97.79%) | 1,928,329 (3.89%) | 46,602,419 (93.91%) |
| PC-3 | 61,563,182 | 59,220,992 | 58,041,957 (98.01%) | 2,122,980 (3.58%) | 55,918,977 (94.42%) |
| Type | Transcript Number (percent) | Gene Number (percent) |
|---|---|---|
| COG | 16,917 (8.49%) | 16,890 (29%) |
| GO | 70,127 (35.2%) | 19,197 (9.64%) |
| KEGG | 104,630 (52.52%) | 13,266 (6.66%) |
| Total annotation | 122,847 (61.66%) | 19,937 (10.01%) |
| Total | 199,234 | 58,233 |
| Total | Up | Down | |
|---|---|---|---|
| Total DEGs | 18,639 | 9670 | 8969 |
| Significant DEGs | 2970 | 1539 | 1431 |
| Tanscript/Gene ID | String ID | Degree |
|---|---|---|
| ENSG00000150991 | UBC | 42 |
| ENSG00000141510 | TP53 | 16 |
| ENSG00000104738 | MCM4 | 13 |
| ENSG00000112118 | MCM3 | 12 |
| ENSG00000073111 | MCM2 | 11 |
| ENSG00000100297 | MCM5 | 10 |
| ENSG00000076003 | MCM6 | 9 |
| ENSG00000012048 | BRCA1 | 9 |
| ENSG00000147536 | GINS4 | 8 |
| ENSG00000132646 | PCNA | 8 |
| ENSG00000093009 | CDC45 | 8 |
| ENSG00000097046 | CDC7 | 6 |
| ENSG00000065328 | MCM10 | 6 |
| ENSG00000101003 | GINS1 | 5 |
| ENSG00000076242 | MLH1 | 5 |
| ENSG00000124762 | CDKN1A | 5 |
| ENSG00000171552 | BCL2L1 | 5 |
| ENSG00000187790 | FANCM | 5 |
| ENSG00000055130 | CUL1 | 5 |
| ENSG00000143384 | MCL1 | 5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, S.; Li, S.; Wang, J.; Zhao, L.; Yan, Y.; Cao, Q.; Wu, T.; Liu, L.; Wang, C. Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth. Mar. Drugs 2018, 16, 511. https://doi.org/10.3390/md16120511
Hao S, Li S, Wang J, Zhao L, Yan Y, Cao Q, Wu T, Liu L, Wang C. Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth. Marine Drugs. 2018; 16(12):511. https://doi.org/10.3390/md16120511
Chicago/Turabian StyleHao, Shuai, Shuang Li, Jing Wang, Lei Zhao, Yan Yan, Qi Cao, Tingting Wu, Liyun Liu, and Chengtao Wang. 2018. "Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth" Marine Drugs 16, no. 12: 511. https://doi.org/10.3390/md16120511