Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169
Abstract
:1. Introduction
2. Results
2.1. Taxonomy and Phylogenetic Analysis of the Strain M-157
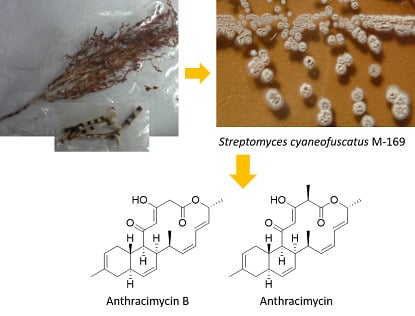
2.2. Isolation and Structural Elucidation of Compounds 1 and 2
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Taxonomic Identification of the Producing Microorganism
4.3. Fermentation of the Producing Microorganism
4.4. Extraction and Isolation
4.5. Antibacterial Activity Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.H.; Nam, S.J.; Locke, J.B.; Kauffman, C.A.; Beatty, D.S.; Paul, L.A.; Fenical, W. Anthracimycin, a Potent Anthrax Antibiotic from a Marine-Derived Actinomycete. Angew. Chem. Int. Ed. 2013, 52, 7822–7824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Head, B.M.; Rubinstein, E.; Meyers, A.F.A. Alternative pre-approved and novel therapies for the treatment of anthrax. BMC Infect. Dis. 2016, 16, 621. [Google Scholar] [CrossRef] [PubMed]
- Hensler, M.E.; Jang, K.H.; Thienphrapa, W.; Vuong, L.; Tran, D.N.; Soubih, E.; Lin, L.; Haste, N.M.; Cunningham, M.L.; Kwan, B.P.; et al. Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus. J. Antibiot. 2014, 67, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Yamashita, T.; Okada, H.; Oishi, N.; Sunagozaka, H.; Nio, K.; Hayashi, T.; Hara, Y.; Asahina, Y.; Yoshida, M.; et al. A Novel mTOR Inhibitor; Anthracimycin for the Treatment of Human Hepatocellular Carcinoma. Anticancer Res. 2017, 37, 3397–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungmann, K.; Jansen, R.; Gerth, K.; Huch, V.; Krug, D.; Fenical, W.; Müller, R. Two of a Kind—The Biosynthetic Pathways of Chlorotonil and Anthracimycin. ACS Chem. Biol. 2015, 10, 2480–2490. [Google Scholar] [CrossRef] [PubMed]
- Alt, S.; Wilkinson, B. Biosynthesis of the Novel Macrolide Antibiotic Anthracimycin. ACS Chem. Biol. 2015, 10, 2468–2479. [Google Scholar] [CrossRef] [PubMed]
- Harunari, E.; Komaki, H.; Igarashi, Y. Biosynthetic origin of anthracimycin: A tricyclic macrolide from Streptomyces sp. J. Antibiot. 2015, 69, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Sirota, F.L.; Goh, F.; Low, K.N.; Yang, L.K.; Crasta, S.C.; Eisenhaber, B.; Eisenhaber, F.; Kanagasundaram, Y.; Ng, S.B. Isolation and Identification of an Anthracimycin Analogue from Nocardiopsis kunsanensis, a Halophile from a Saltern, by Genomic Mining Strategy. J. Genom. 2018, 6, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological Potential of Phylogenetically Diverse Actinobacteria Isolated from Deep-Sea Coral Ecosystems of the Submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Braña, A.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; de la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a New Natural Product with Cytotoxic Activity Produced by Streptomyces sp. M-207 Associated with the Deep-Sea Coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.; de la Cruz, M.; Díaz, C.; Vicente, F.; et al. Branimycins B and C, Antibiotics Produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Vizcaíno, A.; Braña, A.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; Cruz, M.; Díaz, C.; Vicente, F.; Acuña, J.; Reyes, F.; et al. Paulomycin G, a New Natural Product with Cytotoxic Activity against Tumor Cell Lines Produced by Deep-Sea Sediment Derived Micromonospora matsumotoense M-412 from the Avilés Canyon in the Cantabrian Sea. Mar. Drugs 2017, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-López, F.J.; Alcalde, E.; Sarmiento-Vizcaíno, A.; Díaz, C.; Cautain, B.; García, L.A.; Blanco, G.; Reyes, F. New 3-Hydroxyquinaldic Acid Derivatives from Cultures of the Marine Derived Actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs 2018, 16, 371. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.; Crespo, G.; González-Menéndez, V.; Pérez-Moreno, G.; Sánchez-Carrasco, P.; Pérez-Victoria, I.; Ruiz-Pérez, L.M.; González-Pacanowska, D.; Vicente, F.; Genilloud, O.; et al. MDN-0104, an Antiplasmodial Betaine Lipid from Heterospora chenopodii. J. Nat. Prod. 2014, 77, 2118–2123. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Weissbach, U.; Reillo, C.S.; Braña, A.F.; Méndez, C.; Rohr, J.; Salas, J.A. Identification of two genes from Streptomyces argillaceus encoding two glycosyltransferases involved in the transfer of a disaccharide during the biosynthesis of the antitrumor drug mithramycin. J. Bacteriol. 1998, 180, 4929–4937. [Google Scholar] [PubMed]
- Audoin, C.; Bonhomme, D.; Ivanisevic, J.; Cruz, M.; Cautain, B.; Monteiro, M.; Reyes, F.; Rios, L.; Perez, T.; Thomas, O. Balibalosides, an Original Family of Glucosylated Sesterterpenes Produced by the Mediterranean Sponge Oscarella balibaloi. Mar. Drugs 2013, 11, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Martín, J.; de la Cruz, M.; Reyes, F.; Romano, G.; Ianora, A. First identification of marine diatoms with anti-tuberculosis activity. Sci. Rep. 2018, 8, 2284. [Google Scholar] [CrossRef] [PubMed]




| Position | 1 | |||
|---|---|---|---|---|
| δC, Type | δH (J in Hz) | HMBC | COSY | |
| 1 | 165.5, C | |||
| 2 | 46.5, CH2 | 3.50, d (11.3) | 1, 3 | 2b |
| 3.22, d (11.3) | 1, 3 | 2a | ||
| 3 | 184.7, C | |||
| 3-OH | 15.35, br s | |||
| 4 | 102.8, C | 5.96, br s | ||
| 5 | 196.5, C | |||
| 6 | 53.1, CH | 2.63, m | 12, 14, 15, 16 | 7 |
| 7 | 37.3, CH | 1.99, m | 12 | 6, 12 |
| 8 | 31.2, CH2 | 2.42, br d (15.4) | 8b, 9 | |
| 1.51, m | 23 | 8a | ||
| 9 | 120.9, CH | 5.37, m | 7, 11, 12, 23 | 8a |
| 10 | 133.8, C | |||
| 11 | 37.3, CH2 | 2.05, m | 10, 12 | 11b |
| 1.82, br d (16.9) | 10, 23 | 11a | ||
| 12 | 32.7, CH | 2.64, m | 7, 13 | |
| 13 | 133.0, C | 5.73, d (10.0) | 7, 11, 12, 15, 16 | 12, 14 |
| 14 | 124.6, C | 5.54, m | 6, 11, 16 | 13 |
| 15 | 32.9, CH | 1.96, m | 6, 16 | |
| 16 | 45.5, CH | 2.65, m | 14, 17, 24 | 15, 17, 24 |
| 17 | 139.0, CH | 5.40, m | 15, 19, 24 | 16, 18 |
| 18 | 125.9, CH | 5.88, dd (10.8, 10.8) | 15, 19, 20 | 17, 19 |
| 19 | 123.6, CH | 6.48, dd (13.0, 12.7) | 21 | 18, 20 |
| 20 | 131.3, CH | 5.55, m | 18, 21 | 19 |
| 21 | 70.0, CH | 5.57, m | 22 | |
| 22 | 20.7, CH3 | 1.35, d (6.7) | 20, 21 | 21 |
| 23 | 23.4, CH3 | 1.68, s | 9, 10, 11 | |
| 24 | 16.2, CH3 | 0.95, d (6.0) | 15, 16, 17 | 16 |
| Pathogen | Strain | MIC (µg/mL (µM)) | |
|---|---|---|---|
| Anthracimycin (1) | Anthracimycin B (2) | ||
| S. aureus MRSA | MB5393 | <0.03 (<0.076) | 0.125–0.25 (0.33–0.65) |
| S. aureus MSSA | ATCC29213 | <0.03 (<0.076) | 4–8 (10.5–20.9) |
| E. faecium VANS | CL144754 | <0.03 (<0.076) | 0.125–0.25 (0.33–0.65) |
| E. faecalis VANS | CL144492 | <0.03 (<0.076) | 0.25–0.5 (0.65–1.26) |
| E. coli | ATCC25922 | >16 (>40.3) | >16 (>41.8) |
| K. pneumoniae | ATCC700603 | >16 (>40.3) | >16 (>41.8) |
| M. tuberculosis | H37Ra | 1–2 (2.5–5) | >16 (>41.8) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, V.; Martín, J.; Sarmiento-Vizcaíno, A.; De la Cruz, M.; García, L.A.; Blanco, G.; Reyes, F. Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs 2018, 16, 406. https://doi.org/10.3390/md16110406
Rodríguez V, Martín J, Sarmiento-Vizcaíno A, De la Cruz M, García LA, Blanco G, Reyes F. Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169. Marine Drugs. 2018; 16(11):406. https://doi.org/10.3390/md16110406
Chicago/Turabian StyleRodríguez, Víctor, Jesús Martín, Aida Sarmiento-Vizcaíno, Mercedes De la Cruz, Luis A. García, Gloria Blanco, and Fernando Reyes. 2018. "Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169" Marine Drugs 16, no. 11: 406. https://doi.org/10.3390/md16110406