Targeting the Bacterial Protective Armour; Challenges and Novel Strategies in the Treatment of Microbial Biofilm
Abstract
:1. Microbial Biofilms and the Challenges in Infectious Disease
1.1. Methicillin-Resistant Staphylococcus aureus (MRSA)
1.2. Pseudomonas aeruginosa
1.3. Klebsiella pneumoniae
2. The Physiology of Biofilms
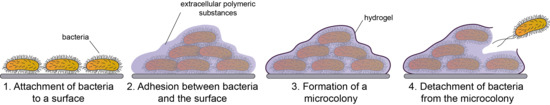
2.1. Definition and the Structure of the Biofilm
2.2. Development of Biofilms
2.3. Mechanism of Antibiotic Resistance in Microbial Biofilms
2.4. Immune Evasion of Biofilms
3. Guideline for Management of Biofilm Associated Infection
4. Diagnosis of Biofilm Mediated Infections
4.1. Sonication
4.2. Polymerase Chain Reaction (PCR)
4.3. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
4.4. Fluorescence In Situ Hybridization (FISH)
4.5. Microscopy
5. The Potency of Existing Therapies against Microbial Biofilm
6. Promising Novel Therapies for Prevention and Treatment of Biofilm Associated Infections
6.1. Nanoparticles
6.2. Diterpenoids
6.3. Biomacromolecules
6.4. Honey
6.5. Antimicrobial Peptides
6.6. Antimicrobial Polymer
7. Summary and Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. Apmis 2013, 121, 1–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Hu, X.; Hu, Z.; Tang, Z.; Wu, H.; Chen, L. Morbidity and mortality of nosocomial infection after cardiovascular surgery: A report of 1606 cases. Curr. Med. Sci. 2018, 38, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Barrasa-Villar, J.I.; Aibar-Remón, C.; Prieto-Andrés, P.; Mareca-Doñate, R.; Moliner-Lahoz, J. Impact on morbidity, mortality and length of stay of hospital acquired infections by resistant microorganisms. Clin. Infect. Dis. 2017, 65, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Edwards, J.R.; Richards, C.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, I.; Schofield, C.; Everett, M.; O’Neill, A.; Miller, K.; Wilcox, M.; Frere, J.M.; Dawson, M.; Czaplewski, L.; Urleb, U.; et al. Treatment of health-care-associated infections caused by Gram-negative bacteria: A consensus statement. Lancet Infect. Dis. 2008, 8, 133–139. [Google Scholar] [CrossRef]
- Allegranzi, B.; Nejad, S.B.; Combescure, C.; Graafmans, W.; Attar, H.; Donaldson, L.; Pittet, D. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet 2011, 377, 228–241. [Google Scholar] [CrossRef]
- Herman-Bausier, P.; Dufrene, Y.F. Force matters in hospital-acquired infections. Science 2018, 359, 1464–1465. [Google Scholar] [CrossRef] [PubMed]
- Mataraci, E.; Dosler, S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 2012, 56, 6366–6371. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Suleman, L.; Donelli, G. Healthcare-Associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Al-Talib, H.; Yean, C.; Al-Jashamy, K.; Hasan, H. Methicillin-resistant Staphylococcus aureus nosocomial infection trends in Hospital Universiti Sains Malaysia during 2002–2007. Ann. Saudi Med. 2010, 30, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Macmorran, E.; Harch, S.; Athan, E.; Lane, S.; Tong, S.; Crawford, L.; Krishnaswamy, S.; Hewagama, S. The rise of methicillin resistant Staphylococcus aureus: Now the dominant cause of skin and soft tissue infection in Central Australia. Epidemiol. Infect. 2017, 145, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef] [PubMed]
- Malani, P.N. National burden of invasive methicillin-resistant Staphylococcus aureus infection. JAMA 2014, 311, 1438–1439. [Google Scholar] [CrossRef] [PubMed]
- Sisirak, M.; Zvizdic, A.; Hukic, M. Methicillin-resistant Staphylococcus aureus (MRSA) as a cause of nosocomial wound infections. Bosn. J. Basic Med. Sci. 2010, 10, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, N.F.; Firdessa, R.; Good, L. Bactericidal effects of polyhexamethylene biguanide against intracellualar Staphylococcus aureus EMRSA-15 and USA 300. J. Antimicrob. Chemother. 2016, 71, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, G. Diagnosis and management of complicated intra-abdominal infection in adults and children: Guidelines by the surgical infection society and the infectious diseases society of America. Chin. J. Infect. Chemother. 2010, 10, 241–247. [Google Scholar] [CrossRef]
- Codjoe, F.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Johansen, H.K.; Moskowitz, S.M.; Ciofu, O.; Pressler, T.; Høiby, N. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J. Cyst. Fibros. 2008, 7, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Criss, A.K.; Katz, B.Z.; Seifert, H.S. Resistance of Neisseria gonorrhoeae to non-oxidative killing by adherent human polymorphonuclear leucocytes. Cell. Microbiol. 2009, 11, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Fiaccadori, E.; Antonucci, E.; Morabito, S.; d’Avolio, A.; Maggiore, U.; Regolisti, G. Colistin Use in Patients With Reduced Kidney Function. Am. J. Kidney Dis. 2016, 68, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Anju, C.P.; Biswas, L.; Anil Kumar, V.; Gopi Mohan, C.; Biswas, R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int. J. Med. Microbiol. 2016, 306, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Juárez, P.; Vilar-Compte, D.; Pérez-Jiménez, C.; Ñamendys-Silva, S.A.; Sandoval-Hernández, S.; Volkow-Fernández, P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int. J. Infect. Dis. 2015, 31, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Golan, Y. Empiric therapy for hospital-acquired, Gram-negative complicated intra-abdominal infection and complicated urinary tract infections: A systematic literature review of current and emerging treatment options. BMC Infect. Dis. 2015, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yayan, J.; Ghebremedhin, B.; Rasche, K. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single university hospital center in Germany over a 10-Year Period. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Mcguffie, B.A.; Vallet-gely, I.; Dove, S.L. σ factor and anti-σ factor that control swarming motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2016, 198, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Chaftari, A.M.; Zakhem, A.E.; Jamal, M.A.; Jiang, Y.; Hachem, R.; Raad, I. The use of minocycline-rifampin coated central venous catheters for exchange of catheters in the setting of Staphylococcus aureus central line associated bloodstream infections. BMC Infect. Dis. 2014, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Surveillance of Antimicrobial Resistance in Europe 2016; Annual report of the European Antimicrobial REsistance Surveillance Network (EARS-Net); European Centre for Disease Prevention and Control: Solna, Sweden, 2017; ISBN 9789294980991.
- Livermore, D.M.; Maya, J.J.; Nordmann, P.; Wang, H.; Woodford, N.; Quinn, J.P. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Otter, J.A.; Doumith, M.; Davies, F.; Mookerjee, S.; Dyakova, E.; Gilchrist, M.; Brannigan, E.T.; Bamford, K.; Galletly, T.; Donaldson, H.; et al. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Singla, S.; Harjai, K.; Chhibber, S. Artificial Klebsiella pneumoniae biofilm model mimicking in vivo system: Altered morphological characteristics and antibiotic resistance. J. Antibiot. 2014, 67, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.Y. The emerging problems of Klebsiella pneumoniae infections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Jendresen, M.D.; Glantz, P.O. Clinical adhesiveness of selected dental materials: An in-vivo study. Acta Odontol. Scand. 1981, 39, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N. A personal history of research on microbial biofilms and biofilm infections. Pathog. Dis. 2014, 70, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, E.; Roe, F.; Bugnicourt, A.; Michael, J.; Heydorn, A.; Molin, S.; Pitts, B.; Stewart, P.S.; Franklin, M.J. Stratified growth in Pseudomonas aeruginosa biofilms stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2004, 70, 6188–6196. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, N.F.; Chong, S.Q.Y.; Edmondson-Brown, K.M.; Ntow-Boahene, W.; Bardiau, M.; Good, L. Bactericidal and anti-biofilm effects of polyhexamethylene Biguanide in models of intracellular and biofilm of Staphylococcus aureus isolated from bovine mastitis. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Czaczyk, K.; Myszka, K. Biosynthesis of extracellular polymeric substances (EPS) and its role in microbial biofilm formation. Pol. J. Environ. Stud. 2007, 16, 799–806. [Google Scholar]
- Izano, E.A.; Amarante, M.A.; Kher, W.B.; Kaplan, J.B. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008, 74, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Billings, N.; Ramirez Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013, 9, e1003526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sehar, S.; Koop, L.; Wong, Y.K.; Ahmed, S.; Siddiqui, K.S.; Manefield, M. Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS ONE 2014, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bos, R.; Van Der Mei, H.C.; Busscher, H.J. Physico-chemistry of initial microbial adhesive interactions-its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–229. [Google Scholar] [CrossRef] [PubMed]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2013, 41, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Poggi, A.; Visai, L.; Ravaioli, S.; Campoccia, D.; Speziale, P.; Arciola, C.R. Extracellular DNA in biofilms. Int. J. Artif. Organs 2011, 34, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Costerton, A.J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 2011, 284, 1318–1322. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.C.; Toole, G.A.O. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- La, B.; Prosser, T.; Taylor, D.; Dix, B.A.; Cleeland, R.O.Y. Method of evaluating effects of antibiotics on bacterial biofilm. Antimicrob. Agents Chemother. 1987, 31, 1502–1506. [Google Scholar]
- Nickel, J.C.; Ruseska, I.; Wright, J.B.; Costerton, J.W. Tobramycin resistance of cells of Pseudomonas aeruginosa growing as biofilm on urinary catheter material. Antimicrob. Agents Chemother. 1985, 27, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Sahore, S.; Kaur, P.; Rani, A.; Ray, P. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog. Dis. 2016, 74, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Joshi-Datar, A.; Lepine, F.; Bauerle, E.; Olakanmi, O.; Beer, K.; McKay, G.; Siehnel, R.; Schafhauser, J.; Wang, Y.; et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011, 334, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Bowler, L.L.; Zhanel, G.G.; Ball, T.B.; Saward, L.L. Mature Pseudomonas aeruginosa biofilms prevail compared to young biofilms in the presence of ceftazidime. Antimicrob. Agents Chemother. 2012, 56, 4976–4979. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.; Horsman, S.R.; Charron-Mazenod, L.; Turnbull, A.L.; Mulcahy, H.; Surette, M.G.; Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of Antibiotic Penetration, Oxygen Limitation. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J.; Williamson, K.S.; Folsom, J.P.; Boegli, L.; James, G.A. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2015, 59, 3838–3847. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.M.; Rowe, S.E.; O’Gara, J.P.; Conlon, B.P. Convergence of Staphylococcus aureus persister and biofilm research: Can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 2016, 12, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Park, M.D.; Otto, M. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front. Microbiol. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Bakaletz, L.O.; Wozniak, D.J. What’s on the outside matters: The role of the extracellular polymeric substance of gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J. Biol. Chem. 2016, 291, 12538–12546. [Google Scholar] [CrossRef] [PubMed]
- Roilides, E.; Simitsopoulou, M.; Katragkou, A.; Walsh, T.J. How Biofilms Evade Host Defenses. In Microbial Biofilms, 2nd ed.; American Society for Microbiology: Washington, DC, USA, 2015; pp. 287–300. ISBN1 978-1-55581-745-9. ISBN2 978-1-55581-746-6. [Google Scholar]
- Leid, J.G.; Shirtliff, M.E.; Costerton, J.W.; Stoodley, P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 2002, 70, 6339–6345. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Tomlin, K.L.; Worthen, G.S.; Poch, R.; Lieber, J.G.; Saavedra, M.T.; Michael, B.; Malcolm, K.C.; Vasil, M.L.; Jerry, A.N.; et al. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 2005, 73, 3693–3701. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef] [PubMed]
- Jesaitis, A.J.; Franklin, M.J.; Berglund, D.; Sasaki, M.; Lord, C.I.; Bleazard, J.B.; Duffy, J.E.; Beyenal, H.; Lewandowski, Z. Compromised host defense on Pseudomonas aeruginosa biofilms: Characterization of neutrophil and biofilm interactions. J. Immunol. 2003, 171, 4329–4339. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.T.; Kharazmi, A.; Lam, K.; Costerton, J.W.; Hoiby, N. Human polymorphonuclear leukocyte response to Pseudomonas aeruginosa grown in biofilms. Infect. Immun. 1990, 58, 2383–2385. [Google Scholar] [PubMed]
- Wu, H.; Moser, C.; Wang, H.Z.; Høiby, N.; Song, Z.J. Strategies for combating bacterial biofilm infections. Int. J. Oral. Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Holá, V.; Imbert, C.; Kirketerp-Møller, K.; et al. ESCMID* guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015, 21, S1–S25. [Google Scholar] [CrossRef] [PubMed]
- Macià, M.D.; del Pozo, J.L.; Díez-Aguilar, M.; Guinea, J. Microbiological diagnosis of biofilm-related infections. Enferm. Infecc. Microbiol. Clin. 2018, 36, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Almani, S.A.; Naseer, A.; Maheshwari, S.K.; Maroof, P. Detection of biofilm-associated implant pathogens in cardiac device infections: High sensitivity of sonication fluid culture even in the presence of antimicrobials. J. Glob. Infect. Dis. 2017, 9, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Mandakhalikar, K.D.; Rahmat, J.N.; Chiong, E.; Neoh, K.G.; Shen, L.; Tambyah, P.A. Extraction and quantification of biofilm bacteria: Method optimized for urinary catheters. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Renz, N.; Cabric, S.; Morgenstern, C.; Schuetz, M.A.; Trampuz, A. Value of PCR in sonication fluid for the diagnosis of orthopedic hardware-associated infections: Has the molecular era arrived? Injury 2018, 49, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Schuurs, T.A.; Koelewijn, R.; Brienen, E.A.T.; Kortbeek, T.; Mank, T.G.; Mulder, B.; Stelma, F.F.; Van Lieshout, L.; Van Hellemond, J.J. Harmonization of PCR-based detection of intestinal pathogens: Experiences from the Dutch external quality assessment scheme on molecular diagnosis of protozoa in stool samples. Clin. Chem. Lab. Med. 2018, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Stavnsbjerg, C.; Frimodt-Møller, N.; Moser, C.; Bjarnsholt, T. Comparison of two commercial broad-range PCR and sequencing assays for identification of bacteria in culture-negative clinical samples. BMC Infect. Dis. 2017, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Dannaoui, E.; Chouaki, T.; Cateau, E.; Malard, O.; Bonfils, P.; Page, C.; Dufour, X.; Cottrel, C.; Erwan, T.; et al. PCR-based detection of Aspergillus fumigatus and absence of azole resistance due to TR34/L98H in a french multicenter cohort of 137 patients with fungal rhinosinusitis. Mycoses 2018, 61, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Tyler, K.L. Molecular Diagnosis of CNS Viral Infections; Elsevier Inc.: New York, NY, USA, 2005; Voluem 76, ISBN 9780128138069. [Google Scholar]
- Bizzini, A.; Greub, G.; Greub, G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 2010, 16, 1614–1619. [Google Scholar] [CrossRef] [PubMed]
- Kliem, M.; Sauer, S. The essence on mass spectrometry based microbial diagnostics. Curr. Opin. Microbiol. 2012, 15, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Gaudreau, A.M.; Labrie, J.; Goetz, C.; Dufour, S.; Jacques, M. Evaluation of MALDI-TOF mass spectrometry for the identification of bacteria growing as biofilms. J. Microbiol. Methods 2018, 145, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Schrøder, S.A.; Eickhardt, S.; Bjarnsholt, T.; Nørgaard, T.; Homøe, P. Morphological evidence of biofilm in chronic obstructive sialadenitis. J. Laryngol. Otol. 2018, 132, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.; Wunder, A.; Auschill, T.M.; Follo, M.; Braun, G.; Hellwig, E.; Arweiler, N.B. The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. J. Med. Microbiol. 2007, 56, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Hardy, L.; Jespers, V.; Dahchour, N.; Mwambarangwe, L.; Musengamana, V.; Vaneechoutte, M.; Crucitti, T. Unravelling the bacterial vaginosis-associated biofilm: A multiplex Gardnerella vaginalis and Atopobium vaginae fluorescence in situ hybridization assay using peptide nucleic acid probes. PLoS ONE 2015, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Johani, K.; Gosbell, I.B.; Jacombs, A.S.W.; Almatroudi, A.; Whiteley, G.S.; Deva, A.K.; Jensen, S.; Vickery, K. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: Combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J. Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Mohmmed, S.A.; Vianna, M.E.; Penny, M.R.; Hilton, S.T.; Mordan, N.; Knowles, J.C. Confocal laser scanning, scanning electron, and transmission electron microscopy investigation of Enterococcus faecalis biofilm degradation using passive and active sodium hypochlorite irrigation within a simulated root canal model. Microbiologyopen 2017, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tani, S.; Lepetsos, P.; Stylianakis, A.; Vlamis, J.; Birbas, K.; Kaklamanos, I. Superiority of the sonication method against conventional periprosthetic tissue cultures for diagnosis of prosthetic joint infections. Eur. J. Orthop. Surg. Traumatol. 2018, 28, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Forbes, M.; Greenberg, D.P.; Dice, B.; Burrows, A.; Wackym, P.A.; Kerschner, J.E. Direct Detection of Bacterial Biofilms on the Middle-Ear Mucosa of Children. JAMA 2006, 296, 202–211. [Google Scholar]
- Renz, N.; Feihl, S.; Cabric, S.; Trampuz, A. Performance of automated multiplex PCR using sonication fluid for diagnosis of periprosthetic joint infection: A prospective cohort. Infection 2017, 45, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Boase, S.; Foreman, A.; Cleland, E.; Tan, L.; Melton-Kreft, R.; Pant, H.; Hu, F.Z.; Ehrlich, G.D.; Wormald, P.J. The microbiome of chronic rhinosinusitis: Culture, molecular diagnostics and biofilm detection. BMC Infect. Dis. 2013, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Ciofu, O.; Molin, S.; Givskov, M.; Høiby, N. Applying insights from biofilm biology to drug development-can a new approach be developed? Nat. Rev. Drug Discov. 2013, 12, 791–808. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Máiz, L.; Escribano, A.; Olveira, C.; Oliver, A.; Asensio, O.; Gartner, S.; Roma, E.; Quintana-Gallego, E.; Salcedo, A.; et al. Spanish Consensus on the Prevention and Treatment of Pseudomonas aeruginosa Bronchial Infections in Cystic Fibrosis Patients. Arch. Bronconeumol. 2015, 51, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Haworth, C.S.; Foweraker, J.E.; Wilkinson, P.; Kenyon, R.F.; Bilton, D. Inhaled colistin in patients with bronchiectasis and chronic Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 2014, 189, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Welte, T.; Polverino, E.; De Soyza, A.; Greville, H.; O’Donnell, A.; Alder, J.; Reimnitz, P.; Hampel, B. Ciprofloxacin dry powder for inhalation in non-cystic fibrosis bronchiectasis: A phase II randomised study. Eur. Respir. J. 2013, 41, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, W.F.; Fawaz, S.A.; Rabie, H.; Hamdy, T.A.; Shokry, Y.A. Effect of topical ofloxacin on bacterial biofilms in refractory post-sinus surgery rhino-sinusitis. Eur. Arch. Oto-Rhino-Laryngol. 2015, 272, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, M.; Gil, J.; Treu, R.; Valdes, J.; Davis, S. An in vitro analysis of the effects of various topical antimicrobial agents on methicillin-resistant and methicillin-sensitive strains of Staphylococcus aureus. Ostomy Wound Manag. 2014, 60, 18–28. [Google Scholar]
- Hogan, S.; Zapotoczna, M.; Stevens, N.T.; Humphreys, H.; O’Gara, J.P.; O’Neill, E. In vitro approach for identification of the most effective agents for antimicrobial lock therapy in the treatment of intravascular catheter-related infections caused by Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Moghaddas, A.; Abbasi, M.R.; Gharekhani, A.; Dashti-Khavidaki, S.; Razeghi, E.; Jafari, A.; Khalili, H. Prevention of hemodialysis catheter-related blood stream infections using a cotrimoxazole-lock technique. Future Microbiol. 2015, 10, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Mataraci Kara, E.; Ozbek Celik, B. Investigation of the effects of various antibiotics against Klebsiella pneumoniae biofilms on in vitro catheter model. J. Chemother. 2018, 30, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, J.; Bode-Boger, S.M.; Tröger, U.; Martens-Lobenhoffer, J.; Mulrooney, T.; Mittelstädt, H.; Russlies, M.; Kirchner, R.; Knobloch, J.K.M. Successful treatment of extensively drug-resistant Pseudomonas aeruginosa osteomyelitis using a colistin- and tobramycin-impregnated PMMA spacer. Int. J. Antimicrob. Agents 2014, 44, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Cancienne, J.M.; Tyrrell Burrus, M.; Weiss, D.B.; Yarboro, S.R. Applications of Local Antibiotics in Orthopedic Trauma. Orthop. Clin. N. Am. 2015, 46, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Subbenaik, S.C. Plant Nanotechnology: Principles and Practices; Khodakovskaya, C.K.S.K.V., Ed.; Springer International Publishing: Basel, Switzerland, 2016; ISBN 9783319421544. [Google Scholar]
- Yadav, N.; Dubey, A.; Shukla, S.; Saini, C.P.; Gupta, G.; Priyadarshini, R.; Lochab, B. Graphene oxide-coated surface: Inhibition of bacterial biofilm formation due to specific surface-interface interactions. ACS Omega 2017, 2, 3070–3082. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, M.J.; Fromm, K.M.; Akbar Ashkarran, A.; Jimenez de Aberasturi, D.; De Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aditya, A.; Chattopadhyay, S.; Jha, D.; Gautam, H.K.; Maiti, S.; Ganguli, M. Zinc Oxide nanoparticles dispersed in ionic liquids show high antimicrobial efficacy to skin-specific bacteria. ACS Appl. Mater. Interfaces 2018, 10, 15401–15411. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Coenye, T. The Role of Reactive Oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Chindera, K.; Mahato, M.; Kumar Sharma, A.; Horsley, H.; Kloc-Muniak, K.; Kamaruzzaman, N.F.; Kumar, S.; McFarlane, A.; Stach, J.; Bentin, T.; et al. The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes. Sci. Rep. 2016, 6, 23121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, H.; Cao, H.; Zhao, Y.; Zhu, C.; Cheng, T.; Wang, Q.; Peng, X.; Cheng, M.; Wang, J.; Jin, G.; et al. In vitro and in vivo anti-biofilm effects of silver nanoparticles immobilized on titanium. Biomaterials 2014, 35, 9114–9125. [Google Scholar] [CrossRef] [PubMed]
- Abdulkareem, E.H.; Memarzadeh, K.; Allaker, R.P.; Huang, J.; Pratten, J.; Spratt, D. Anti-biofilm activity of zinc oxide and hydroxyapatite nanoparticles as dental implant coating materials. J. Dent. 2015, 43, 1462–1469. [Google Scholar] [CrossRef] [PubMed]
- Allaker, R.P. Critical review in oral biology & medicine: The use of nanoparticles to control oral biofilm formation. J. Dent. Res. 2010, 89, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Kwon, D.N.; Kim, J.H. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res. Lett. 2014, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jalvo, B.; Faraldos, M.; Bahamonde, A.; Rosal, R. Antimicrobial and antibiofilm efficacy of self-cleaning surfaces functionalized by TiO2 photocatalytic nanoparticles against Staphylococcus aureus and Pseudomonas putida. J. Hazard. Mater. 2017, 340, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Shakibaie, M.; Forootanfar, H.; Golkari, Y.; Mohammadi-Khorsand, T.; Shakibaie, M.R. Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. J. Trace Elem. Med. Biol. 2015, 29, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Tchimene, M.K.; Okunji, C.O.; Iwu, M.M.; Kuete, V. Monoterpenes and Related Compounds from the Medicinal Plants of Africa. In Medicinal Plant Research in Africa; Elsevier: New York, NY, USA, 2013; pp. 261–300. ISBN 9780124059276. [Google Scholar]
- Manner, S.; Vahermo, M.; Skogman, M.E.; Krogerus, S.; Vuorela, P.M.; Yli-Kauhaluoma, J.; Fallarero, A.; Moreira, V.M. New derivatives of dehydroabietic acid target planktonic and biofilm bacteria in Staphylococcus aureus and effectively disrupt bacterial membrane integrity. Eur. J. Med. Chem. 2015, 102, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Kuźma, Ł.; Rózalski, M.; Walencka, E.; Rózalska, B.; Wysokińska, H. Antimicrobial activity of diterpenoids from hairy roots of Salvia sclarea L.: Salvipisone as a potential anti-biofilm agent active against antibiotic resistant Staphylococci. Phytomedicine 2007, 14, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, J.; Griesser, H.J.; Textor, M.; Landmann, R.; Khanna, N. Antimicrobial properties of 8-hydroxyserrulat-14-en-19-oic acid for treatment of implant-associated infections. Antimicrob. Agents Chemother. 2013, 57, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.P.K.; Coady, D.J.; Sardon, H.; Yuen, A.; Gao, S.; Lim, S.W.; Liang, Z.C.; Tan, E.W.; Venkataraman, S.; Engler, A.C.; et al. Broad Spectrum Macromolecular Antimicrobials with Biofilm Disruption Capability and In Vivo Efficacy. Adv. Healthc. Mater. 2017, 6, 1–9. [Google Scholar] [CrossRef]
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Goncalves, M.; Delattre, C.; Balestrino, D.; Charbonnel, N.; Elboutachfaiti, R.; Wadouachi, A.; Badel, S.; Bernardi, T.; Michaud, P.; Forestier, C. Anti-biofilm activity: A function of Klebsiella pneumoniae capsular polysaccharide. PLoS ONE 2014, 9, e99995. [Google Scholar] [CrossRef]
- Brian-Jaisson, F.; Molmeret, M.; Fahs, A.; Guentas-Dombrowsky, L.; Culioli, G.; Blache, Y.; Cérantola, S.; Ortalo-Magné, A. Characterization and anti-biofilm activity of extracellular polymeric substances produced by the marine biofilm-forming bacterium pseudoalteromonas ulvae strain TC14. Biofouling 2016, 32, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Augustine, N.; Kumar, P.; Thomas, S. Inhibition of Vibrio cholerae biofilm by AiiA enzyme produced from Bacillus spp. Arch. Microbiol. 2010, 192, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.; McWhorter, A.R.; Chousalkar, K.K. Anti-bacterial and anti-biofilm activity of commercial organic acid products against Salmonella enterica isolates recovered from an egg farm environment. Avian Pathol. 2018, 47, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Francolini, I.; Donelli, G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 2010, 59, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junter, G.A.; Thébault, P.; Lebrun, L. Polysaccharide-based antibiofilm surfaces. Acta Biomater. 2016, 30, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R. Honey as an effective antimicrobial treatment for chronic wounds: Is there a place for it in modern medicine? Chronic Wound Care Manag. Res. 2014, 1, 15–22. [Google Scholar] [CrossRef]
- Simon, A.; Traynor, K.; Santos, K.; Blaser, G.; Bode, U.; Molan, P. Medical honey for wound carestill the latest resort. Evid.-Based Complement. Altern. Med. 2009, 6, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Biglari, B.; Swing, T.; Büchler, A.; Ferbert, T.; Simon, A.; Schmidmaier, G.; Moghaddam, A. Medical honey in professional wound care. Expert Rev. Dermatol. 2013, 8, 51–56. [Google Scholar] [CrossRef]
- Watts, R.; Frehner, E. Evidence Summary: Wound management: Medical-grade honey. Wound Pract. Res. 2017, 25, 117–120. [Google Scholar] [CrossRef]
- Müller, P.; Alber, D.G.; Turnbull, L.; Schlothauer, R.C.; Carter, D.A.; Whitchurch, C.B.; Harry, E.J. Synergism between Medihoney and Rifampicin against Methicillin-Resistant Staphylococcus aureus (MRSA). PLoS ONE 2013, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Cokcetin, N.N.; Lu, J.; Turnbull, L.; Carter, D.A.; Whitchurch, C.B.; Harry, E.J. Rifampicin-manuka honey combinations are superior to other antibiotic-manuka honey combinations in eradicating Staphylococcus aureus biofilms. Front. Microbiol. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lu, J.; Müller, P.; Turnbull, L.; Burke, C.M.; Schlothauer, R.C.; Carter, D.A.; Whitchurch, C.B.; Harry, E.J. Antibiotic-specific differences in the response of Staphylococcus aureus to treatment with antimicrobials combined with manuka honey. Front. Microbiol. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Paramasivan, S.; Drilling, A.J.; Jardeleza, C.; Jervis-Bardy, J.; Vreugde, S.; Wormald, P.J. Methylglyoxal-augmented manuka honey as a topical anti-Staphylococcus aureus biofilm agent: Safety and efficacy in an in vivo model. Int. Forum Allergy Rhinol. 2014, 4, 187–195. [Google Scholar] [CrossRef] [PubMed]
- El-Kased, R.F.; Amer, R.I.; Attia, D.; Elmazar, M.M. Honey-based hydrogel: In vitro and comparative in vivo evaluation for burn wound healing. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Lu, J.; Carter, D.A.; Turnbull, L.; Rosendale, D.; Hedderley, D.; Stephens, J.; Gannabathula, S.; Steinhorn, G.; Schlothauer, R.C.; Whitchurch, C.B.; et al. The Effect of New Zealand Kanuka, Manuka and Clover The effect of New Zealand kanuka, manuka and clover honeys on bacterial growth dynamics and cellular morphology varies according to the species. PLoS ONE 2013, 8, e55898. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A. Comparative Antibacterial and Antibiofilm Activities of Manuka Honey and Egyptian Clover Honey. Asian J. Appl. Sci. 2014, 2, 110–115. [Google Scholar]
- Majtan, J.; Bohova, J.; Horniackova, M.; Klaudiny, J.; Majtan, V. Anti-biofilm effects of honey against wound pathogens proteus mirabilis and enterobacter cloacae. Phyther. Res. 2014, 28, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Campeau, M.E.M.; Patel, R.; Campeau, M.E.M.; Patel, R. Antibiofilm Activity of Manuka Honey in Combination with Antibiotics. Int. J. Bacteriol. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-L.; Yu, Q.-H.; Chen, S.-K.; Wang, Y.-H. In-vitro Activity of Honey and Topical Silver in Wound Care Management. Drug Res. 2015, 65, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Sojka, M.; Valachova, I.; Bucekova, M.; Majtan, J. Antibiofilm efficacy of honey and bee-derived defensin-1 on multispecies wound biofilm. J. Med. Microbiol. 2016, 65, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halstead, F.D.; Webber, M.A.; Oppenheim, B.A. Use of an engineered honey to eradicate preformed biofilms of important wound pathogens: An in vitro study. J. Wound Care 2017, 26, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, M.; Karpiński, P.; Pituch, H.; van Belkum, A.; Obuch-Woszczatyński, P. Antimicrobial effects of Manuka honey on in vitro biofilm formation by Clostridium difficile. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Emineke, S.; Cooper, A.J.; Fouch, S.; Birch, B.R.; Lwaleed, B.A. Diluted honey inhibits biofilm formation: Potential application in urinary catheter management? J. Clin. Pathol. 2017, 70, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Turnbull, L.; Burke, C.M.; Liu, M.; Carter, D.A.; Schlothauer, R.C.; Whitchurch, C.B.; Harry, E.J. Manuka-type honeys can eradicate biofilms produced by Staphylococcus aureus strains with different biofilm-forming abilities. PeerJ 2014, 2, e326. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, O.; Ugur, A.; Isiloglu, M.; Ozcan, F. Evaluatıon of the Antıbacterıal, Antıbıofılm, Antıoxıdant, and Cytotoxıc Effects of Some Turkısh Honeys. In Proceedings of the XXXXIII International Apicultural Congres, Kyiv, Ukraine, 29 September–4 October 2013. [Google Scholar]
- Aissat, S.; Ahmed, M.; Djebli, N. Propolis-Sahara honeys preparation exhibits antibacterial and anti-biofilm activity against bacterial biofilms formed on urinary catheters. Asian Pac. J. Trop. Dis. 2016, 6, 873–877. [Google Scholar] [CrossRef]
- da Silva, C.I.; Aazza, S.; Faleiro, M.L.; Miguel, M.D.G.; Neto, L. Propiedades antibacterianas, anti-biofilm, anti-inflamatorias y de inhibición de la virulencia de mieles portuguesas. J. Apic. Res. 2016, 55, 292–304. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Lehrer, R.I. Antibiotic peptides from higher eukaryotes: Biology and applications. Mol. Med. Today 1999, 5, 292–297. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta -Biomembr. 2016, 1858, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Hancock, R.E.W.W. Anti-biofilm peptides: Potential as broad-spectrum agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chou, S.; Xu, L.; Zhu, X.; Dong, N.; Shan, A.; Chen, Z. High specific selectivity and Membrane-Active Mechanism of the synthetic centrosymmetric α-helical peptides with Gly-Gly pairs. Sci. Rep. 2015, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente-Núñez, C.; Cardoso, M.H.; De Souza Cândido, E.; Franco, O.L.; Hancock, R.E.W. Synthetic antibiofilm peptides. Biochim. Biophys. Acta -Biomembr. 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Nadres, E.T.; Kuroda, K. Cationic amphiphilic polymers with antimicrobial activity for oral care applications: Eradication of S. mutans biofilm. Biomacromolecules 2017, 181, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Bonilla, A.; Fernández-García, M. Polymeric materials with antimicrobial activity. Prog. Polym. Sci. 2012, 37, 281–339. [Google Scholar] [CrossRef]
- Li, F.; Chai, Z.G.; Sun, M.N.; Wang, F.; Ma, S.; Zhang, L.; Fang, M.; Chen, J.H. Anti-biofilm effect of dental adhesive with cationic monomer. J. Dent. Res. 2009, 88, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Vishwakarma, A.; Li, Z.; Miyoshi, T.; Barton, H.; Joy, A. Modification of a conventional polyurethane composition provides significant anti-biofilm activity against Escherichia coli. Polym. Chem. 2018, 776. [Google Scholar] [CrossRef]
- Kaehn, K. Polihexanide: A safe and highly effective biocide. Skin Pharmacol. Physiol. 2010, 23, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, N.F.; Pina, M.D.F.; Chivu, A.; Good, L. Polyhexamethylene biguanide and nadifloxacin self-assembled nanoparticles: Antimicrobial effects against intracellular methicillin-resistant Staphylococcus aureus. Polymers (Basel) 2018, 10, 521. [Google Scholar] [CrossRef]
- Lefebvre, E.; Lembre, P.; Picard, J.; El-Guermah, L.; Seyer, D.; Larreta Garde, V. Ephemeral biogels to control anti-biofilm agent delivery: From conception to the construction of an active dressing. Mater. Sci. Eng. C 2018, 82, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Cutting, K.F. Addressing the challenge of wound cleansing in the modern era. Br. J. Nurs. 2010, 19, S24–S29. [Google Scholar] [CrossRef] [PubMed]




| Physiology of Biofilm | Mechanism of Antimicrobial Resistance | References |
|---|---|---|
| Component of the biofilm matrix | ||
|
| [52,54,56,57] |
|
| [42] |
|
| [58] |
|
| [59] |
| Nutritional factors | ||
|
| [60,61,62] |
|
| [57] |
| Physiology of bacteria | ||
|
| [63] |
| Steps | Detail Action |
|---|---|
| Biofilm diagnosis |
|
| Reporting of biofilm associated infections | Biofilm associated infections may be reported using descriptive terms.
|
Treatment of biofilm related infections
|
|
|
|
|
|
| Monitoring |
|
| Standard care |
|
| Biofilm Mediated Infections | Bacterial Species | Techniques | References |
|---|---|---|---|
| Chronic otitis media | S. pneumoniae, M. catarrhalis, P. aeruginosa, S. aureus, and K. pneumoniae | PCR and SEM | [92] |
| Periprosthetic Infection | Coagulase-negative Staphylococci, Propionibacterium spp., Streptococci and Enterococci | Sonication & PCR | [93] |
| Chronic wound | P. aeruginosa and S. aureus | TEM & FISH | [93] |
| Catheter-associated infection | E. coli | Sonication & SEM | [76] |
| Chronic rhinosinusitis | S. aureus and P. acnes | FISH | [94] |
| Cystic fibrosis | P. aeruginosa | FISH and light microscope | [93] |
| Bacteria | Biofilm Site of Infection | Antibiotic Regimen | Duration | Route of Administration | References |
|---|---|---|---|---|---|
| P. aeruginosa | Lung infection in cystic fibrosis (CF) | 0.5–2 MU colistin, twice daily | Continuous | Inhalation | [96] |
| 300 mg tobramycin, twice daily | 28 days on/off cycles | ||||
| 75 mg aztreonam, three times daily | 28 days on/off cycles | ||||
| 32.5 mg or 65 mg ciprofloxacin, once daily | 28 days | ||||
| Lung infection in non-CF bronchiestasis | 1 MU colistin, twice daily | Continuous | Inhalation | [97] | |
| 32.5 mg ciprofloxacin, twice daily | 28 days | Inhalation | [98] | ||
| P. aeruginosa and/or S. aureus | Rhinosinusitis | 3 drops ofloxacin 0.3%, three times daily | 28 days | Nasal drops | [99] |
| S. aureus | Wounds | Mupirocin 2% ointment | - | Cutaneous | [100] |
| S. aureus | Catheters | 50 mg/mL daptomycin | 24 h | Catheter lumen | [101] |
| 10 mg/mL tigecycline | |||||
| 10 mg/mL rifampicin | |||||
| 10 mg/mL cotrimoxazole + 2500 U/mL heparin | 12–24 h | Catheter lumen | [102] | ||
| Minocycline-rifampin | - | Coating | [28] | ||
| K. pneumoniae | Catheters | doripenem and tobramycin | - | Catheter lumen | [103] |
| P. aeruginosa | Orthopedic procedures | 1 g tobramycin + 12 or 24 MU colistin + 40 g polymethylmethacrylate | - | Intraoperative (PMMA beads) | [104] |
| S. aureus | Orthopedic procedures | 40 mg/mL tobramycin + 1 g vancomycin + 10 mL packet of calcium sulfate | - | Intraoperative (calcium sulfate beads) | [104] |
| 2 mg/mL gentamicin aqueous solution | - | Intraoperative (injection) | [105] |
| Type of Nanoparticles | Microbial Biofilm | References |
|---|---|---|
| Silver (immobilized on titanium) | S. intermedius | [112] |
| Silver | P. aeruginosa, Shigella flexneri, S. aureus and S. pneumonia | [115] |
| Titanium dioxide | S. aureus and P. putida | [116] |
| Selenium and selenium dioxide | S. aureus, P. aeruginosa and Proteus mirabilis | [117] |
| Zinc oxide and combination of zinc oxide and hydroxyapatite | Streptococcus sp. | [113] |
| Graphene oxide | E. coli and S. aureus | [107] |
| Type of Diterpenoids | Microbial Biofilm | Mechanism of Action (Hypothetical) | References |
|---|---|---|---|
Abietane (natural)
| S. aureus, S. epidermidis | NA | [14] |
Abietane (synthetic)
| S. aureus, S. epidermidis | Bacterial membrane or the peptidoglycan (PG) layer | [81] |
Diterpene
| S. aureus (methicillin-susceptible and methicillin-resistant), S. epidermidis | Membranolytic properties as well as a general inhibition of macromolecular biosynthesis | [83] |
| Type of Macromolecule | Microbial Biofilm | Mechanism of Action (Hypothetical) | References |
|---|---|---|---|
| Polyether ether ketone–octafluoropentyl methacrylate surface | - | Reduced protein adsorption | [91] |
| AHL lactonase (AiiA), a metallo-beta-lactamase produced by Bacillus spp. | Vibrio cholerae | blocks quorum sensing in Gram-negative bacteria by hydrolyzing N-acyl-homoserine lactones (AHLs) | [89] |
| Chitosan-based surface coating | S. aureus, S. epidermidis | anti-adhesive and bactericidal via contact membrane disruption | [92] |
| K. pneumoniae capsular polysaccharide | S. aureus, S. epidermidis, E. coli | NA | [87] |
| Commercially available organic acid water additives | Salmonella Typhimurium biofilms | Interference to intracellular pH homeostasis, membrane structure, osmolality and macromolecule synthesis | [90] |
| Synthetic PDMEA MeI polymers | S. epidermidis, S. aureus, E. coli and P. aeruginosa, Candida albicans | Membrane disruption | [84] |
| Type of Honey | Microbial Biofilm | Source |
|---|---|---|
| Manuka |
| [141,142,143,144,145,146,147,148] |
| Clover |
| [141,149] |
| Pumpkin |
| [150] |
| Chestnut and thyme |
| [150] |
| Euphorbia |
| [150,151] |
| Chaste |
| [150] |
| Multifloral |
| [150] |
| Eucalyptus |
| [150] |
| Honeydew |
| [145,150] |
| Lavender, strawberry and citrus |
| [152] |
| Sidr |
| [151] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamaruzzaman, N.F.; Tan, L.P.; Mat Yazid, K.A.; Saeed, S.I.; Hamdan, R.H.; Choong, S.S.; Wong, W.K.; Chivu, A.; Gibson, A.J. Targeting the Bacterial Protective Armour; Challenges and Novel Strategies in the Treatment of Microbial Biofilm. Materials 2018, 11, 1705. https://doi.org/10.3390/ma11091705
Kamaruzzaman NF, Tan LP, Mat Yazid KA, Saeed SI, Hamdan RH, Choong SS, Wong WK, Chivu A, Gibson AJ. Targeting the Bacterial Protective Armour; Challenges and Novel Strategies in the Treatment of Microbial Biofilm. Materials. 2018; 11(9):1705. https://doi.org/10.3390/ma11091705
Chicago/Turabian StyleKamaruzzaman, Nor Fadhilah, Li Peng Tan, Khairun Anisa Mat Yazid, Shamsaldeen Ibrahim Saeed, Ruhil Hayati Hamdan, Siew Shean Choong, Weng Kin Wong, Alexandru Chivu, and Amanda Jane Gibson. 2018. "Targeting the Bacterial Protective Armour; Challenges and Novel Strategies in the Treatment of Microbial Biofilm" Materials 11, no. 9: 1705. https://doi.org/10.3390/ma11091705