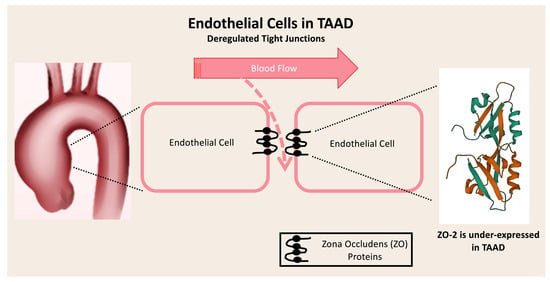
Transcriptomic Analysis of Tight Junction Proteins Demonstrates the Aberrant Expression and Function of Zona Occludens 2 (ZO-2) Protein in Stanford Type A Aortic Dissection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Assessment of the Transcriptomic Profile of the TJPs in Stanford Type A TAAD
2.2. Validation of DEGs as Prognostic Markers for Patients with TAAD
2.3. Nucleotide Sequential Analysis (NSA) of DEGs and Construction of Their Interactome
2.4. Gene Set Enrichment Analysis (GSEA) Regarding the Biological Functions and Regulating miRNAs
2.5. Evaluation of the Identified DEGs Expression Profile in Aneurysmal Aortic Tissue without Dissection
2.6. Statistical Analysis
3. Results
3.1. Assessment of the Transcriptomic Profile of the TJPs in Stanford Type A TAAD
3.2. Validation of DEGs as Prognostic Markers for Patients with TAAD
3.3. Nucleotide Sequential Analysis (NSA) of DEGs and Construction of Their Interactome
3.4. Gene Set Enrichment Analysis (GSEA) Regarding the Biological Functions and Regulating miRNAs
3.5. Evaluation of the Identified DEGs Expression Profile in Aneurysmal Aortic Tissue without Dissection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malashicheva, A.; Kostina, D.; Kostina, A.; Irtyuga, O.; Voronkina, I.; Smagina, L.; Ignatieva, E.; Gavriliuk, N.; Uspensky, V.; Moiseeva, O.; et al. Phenotypic and Functional Changes of Endothelial and Smooth Muscle Cells in Thoracic Aortic Aneurysms. Int. J. Vasc. Med. 2016, 2016, 3107879. [Google Scholar] [CrossRef]
- Elendu, C.; Amaechi, D.C.M.; Elendu, T.C.B.; Ibhiedu, J.O.M.; Torubiri, A.O.M.; Okoye, O.K.M. Comprehensive review of aortic aneurysms, dissections, and cardiovascular complications in connective tissue disorders. Medicine 2023, 102, e36499. [Google Scholar] [CrossRef] [PubMed]
- Naito, S.; Petersen, J.; Sequeira-Gross, T.; Neumann, N.; Escobar, J.D.; Zeller, T.; Reichenspurner, H.; Girdauskas, E. Bicuspid aortopathy—molecular involvement of microRNAs and MMP-TIMP. Biomarkers 2020, 25, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Forte, A.; Della Corte, A.; Grossi, M.; Bancone, C.; Provenzano, R.; Finicelli, M.; De Feo, M.; De Santo, L.S.; Nappi, G.; Cotrufo, M.; et al. Early cell changes and TGFβ pathway alterations in the aortopathy associated with bicuspid aortic valve stenosis. Clin. Sci. 2013, 124, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Kjellqvist, S.; Maleki, S.; Olsson, T.; Chwastyniak, M.; Branca, R.M.M.; Lehtiö, J.; Pinet, F.; Franco-Cereceda, A.; Eriksson, P. A combined proteomic and transcriptomic approach shows diverging molecular mechanisms in thoracic aortic aneurysm development in patients with tricuspid- and bicuspid aortic valve. Mol. Cell. Proteom. 2013, 12, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Wågsäter, D.; Paloschi, V.; Jackson, V.; Petrini, J.; Kurtovic, S.; Maleki, S.; Eriksson, M.J.; Caidahl, K.; Hamsten, A.; et al. Unraveling divergent gene expression profiles in bicuspid and tricuspid aortic valve patients with thoracic aortic dilatation: The ASAP study. Mol. Med. 2011, 17, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yue, J.; Yin, L.; Chen, J.; Chen, Y.; Hu, L.; Shen, J.; Yu, N.; Gong, Y.; Liu, Z. Comprehensive bioinformatics analysis revealed potential key genes and pathways underlying abdominal aortic aneurysm. Comput. Struct. Biotechnol. J. 2023, 21, 5423–5433. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Lilly, B. Notch signaling governs phenotypic modulation of smooth muscle cells. Vasc. Pharmacol. 2014, 63, 88–96. [Google Scholar] [CrossRef]
- Millar, J.K.; Salmon, M.; Nasser, E.; Malik, S.; Kolli, P.; Lu, G.; Pinteaux, E.; Hawkins, R.B.; Ailawadi, G. Endothelial to mesenchymal transition in the interleukin-1 pathway during aortic aneurysm formation. J. Thorac. Cardiovasc. Surg. 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell 2009, 16, 209–221. [Google Scholar] [CrossRef]
- Yang, K.; Cui, S.; Wang, J.; Xu, T.; Du, H.; Yue, H.; Ye, H.; Guo, J.; Zhang, J.; Li, P.; et al. Early Progression of Abdominal Aortic Aneurysm is Decelerated by Improved Endothelial Barrier Function via ALDH2-LIN28B-ELK3 Signaling. Adv. Sci. 2023, 10, e2302231. [Google Scholar] [CrossRef]
- Manzoni, C.; A Kia, D.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; A Lewis, P.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Briefings Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Kimura, N.; Futamura, K.; Arakawa, M.; Okada, N.; Emrich, F.; Okamura, H.; Sato, T.; Shudo, Y.; Koyano, T.K.; Yamaguchi, A.; et al. Gene expression profiling of acute type A aortic dissection combined with in vitro assessment. Eur. J. Cardio-Thoracic Surg. 2017, 52, 810–817. [Google Scholar] [CrossRef]
- Pan, S.; Wu, D.; Teschendorff, A.E.; Hong, T.; Wang, L.; Qian, M.; Wang, C.; Wang, X. JAK2-centered interactome hotspot identified by an integrative network algorithm in acute Stanford type A aortic dissection. PLoS ONE 2014, 9, e89406. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, Y.; Zhu, X.; Tang, X.; Wang, Y.; Wang, J.; Xu, C.; Wang, D.; Du, J.; Zhou, Q. Exaggerated Autophagy in Stanford Type A Aortic Dissection: A Transcriptome Pilot Analysis of Human Ascending Aortic Tissues. Genes 2020, 11, 1187. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.W.; Hosmer, T.; Le Cessie, S.; Lemeshow, S. A comparison of goodness-of-fit tests for the logistic regression model. Stat. Med. 1997, 16, 965–980. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Takai, D.; Jones, P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 2002, 99, 3740–3745. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- E Magouliotis, D.; Fergadi, M.P.; Christodoulidis, G.; A Svokos, A.; A Svokos, K.; Bareka, M.; Athanasiou, T. In-depth bioinformatic study of the cadherin 5 interactome in patients with thoracic aortic aneurysm unveils 8 novel biomarkers. Eur. J. Cardio-Thoracic Surg. 2021, 61, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, C.; Yao, F.; Ding, Q.; Liu, H.; Luo, C.; Wang, D.; Huang, J.; Li, Z.; Shen, Y.; et al. Targeting endothelial tight junctions to predict and protect thoracic aortic aneurysm and dissection. Eur. Hear. J. 2023, 44, 1248–1261. [Google Scholar] [CrossRef]
- Hernández-Guzmán, C.; Gallego-Gutiérrez, H.; Chávez-Munguía, B.; Martín-Tapia, D.; González-Mariscal, L. Zonula occludens 2 and Cell-Cell Contacts Are Required for Normal Nuclear Shape in Epithelia. Cells 2021, 10, 2568. [Google Scholar] [CrossRef]
- Chen, A.; Wu, W.; Lin, S.; Xie, L. Bioinformatic Exploration of Hub Genes and Potential Therapeutic Drugs for Endothelial Dysfunction in Hypoxic Pulmonary Hypertension. Comput. Math. Methods Med. 2022, 2022, 3677532. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, N.; Du, Y. Comprehensive bioinformatics analysis reveals common potential mechanisms, progression markers, and immune cells of coronary virus disease 2019 and atrial fibrillation. Front. Cardiovasc. Med. 2022, 9, 1027026. [Google Scholar] [CrossRef]
- Ahmed, F.F.; Das, A.D.; Sumi, M.J.; Islam, Z.; Rahman, S.; Rashid, H.; Alyami, S.A.; Alotaibi, N.; Azad, A.K.M.; Moni, M.A. Identification of genetic biomarkers, drug targets and agents for respiratory diseases utilising integrated bioinformatics approaches. Sci. Rep. 2023, 13, 19072. [Google Scholar] [CrossRef]





| Gene Symbol | Gene Description |
|---|---|
| OCLN | Occludin |
| CLDN1 | Claudin 1 |
| CLDN2 | Claudin 2 |
| CLDN3 | Claudin 3 |
| CLDN4 | Claudin 4 |
| CLDN5 | Claudin 5 |
| CLDN6 | Claudin 6 |
| CLDN7 | Claudin 7 |
| CLDN8 | Claudin 8 |
| CLDN9 | Claudin 9 |
| CLDN10 | Claudin 10 |
| CLDN11 | Claudin 11 |
| CLDN12 | Claudin 12 |
| CLDN14 | Claudin 14 |
| CLDN15 | Claudin 15 |
| CLDN16 | Claudin 16 |
| CLDN17 | Claudin 17 |
| CLDN18 | Claudin 18 |
| CLDN19 | Claudin 19 |
| CLDN20 | Claudin 20 |
| CLDN22 | Claudin 22 |
| CLDN23 | Claudin 23 |
| CLDN24 | Claudin 24 |
| ZO-1 | Zona Occludens 1 |
| ZO-2 | Zona Occludens 2 |
| ZO-3 | Zona Occludens 3 |
| MAGI-1 | Membrane-Associated Guanylate Kinase, WW And PDZ Domain Containing 1 |
| MAGI-2 | Membrane-Associated Guanylate Kinase, WW And PDZ Domain Containing 2 |
| MAGI-3 | Membrane-Associated Guanylate Kinase, WW And PDZ Domain Containing 3 |
| PAR-3 | Par-3 Family Cell Polarity Regulator |
| PAR-6 | Par-6 Family Cell Polarity Regulator |
| ID | Name | Adjusted p Value | |
|---|---|---|---|
| 1 | GO:0045216 | Cell–cell junction organization | <0.001 |
| 2 | GO:0002934 | Desmosome organization | <0.001 |
| 3 | GO:0007043 | Cell–cell junction assembly | <0.001 |
| 4 | GO:0031581 | Hemidesmosome assembly | <0.001 |
| 5 | GO:0030334 | Regulation of cell migration | <0.001 |
| Regulating miRNA Families | |||
| Name | Adjusted p Value | ||
| 1 | hsa-miR-155-5p | 0.003 | |
| 2 | hsa-miR-1-3p | 0.037 | |
| 3 | hsa-miR-2118-5p | 0.104 | |
| 4 | hsa-miR-4691-3p | 0.142 | |
| 5 | hsa-miR-1229-3p | 0.181 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magouliotis, D.E.; Arjomandi Rad, A.; Kourliouros, A.; Viviano, A.; Koulouroudias, M.; Salmasi, M.Y.; Briasoulis, A.; Triposkiadis, F.; Skoularigis, J.; Athanasiou, T. Transcriptomic Analysis of Tight Junction Proteins Demonstrates the Aberrant Expression and Function of Zona Occludens 2 (ZO-2) Protein in Stanford Type A Aortic Dissection. J. Pers. Med. 2023, 13, 1697. https://doi.org/10.3390/jpm13121697
Magouliotis DE, Arjomandi Rad A, Kourliouros A, Viviano A, Koulouroudias M, Salmasi MY, Briasoulis A, Triposkiadis F, Skoularigis J, Athanasiou T. Transcriptomic Analysis of Tight Junction Proteins Demonstrates the Aberrant Expression and Function of Zona Occludens 2 (ZO-2) Protein in Stanford Type A Aortic Dissection. Journal of Personalized Medicine. 2023; 13(12):1697. https://doi.org/10.3390/jpm13121697
Chicago/Turabian StyleMagouliotis, Dimitrios E., Arian Arjomandi Rad, Antonios Kourliouros, Alessandro Viviano, Marinos Koulouroudias, Mohammad Yousuf Salmasi, Alexandros Briasoulis, Filippos Triposkiadis, John Skoularigis, and Thanos Athanasiou. 2023. "Transcriptomic Analysis of Tight Junction Proteins Demonstrates the Aberrant Expression and Function of Zona Occludens 2 (ZO-2) Protein in Stanford Type A Aortic Dissection" Journal of Personalized Medicine 13, no. 12: 1697. https://doi.org/10.3390/jpm13121697