Aspergillus fumigatus, One Uninucleate Species with Disparate Offspring
Abstract
:1. Introduction
2. Methods
2.1. Strains
2.2. Germination Conditions
2.3. Video Microscopy
2.4. Nucleus and Septum Staining
2.5. Transcriptome
2.6. Epigenetic Inhibitors
2.7. Statistical Analysis
3. Results
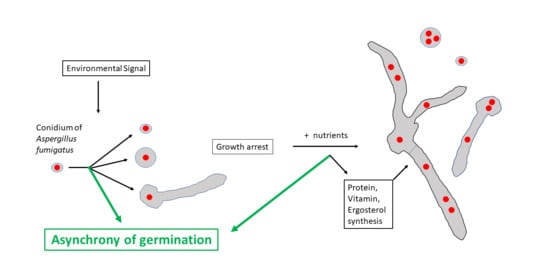
3.1. Characterization of Conidial Germination in A. fumigatus
3.2. Does the Nutritional Environment Modify the Conidial Germination Asynchrony?
3.3. Nuclear Division and Septum Formation in Different Nutritive Environments
3.4. Growth Arrest in G, GA and GAP Media Is Caused by Exhaustion of Nutrients Present in the Conidia
3.5. Transcriptome Analysis
3.6. Trying to Modify the Asynchrony of Conidial Germination in A.fumigatus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lamarre, C.; Sokol, S.; Debeaupuis, J.-P.; Henry, C.; Lacroix, C.; Glaser, P.; Coppée, J.-Y.; François, J.-M.; Latgé, J.-P. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genom. 2008, 9, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osherov, N. Conidial germination in Aspergillus fumigatus. In Aspergillus Fumigatus and Aspergillosis; Latgé, J.-P., Steinbach, W.J., Eds.; ASM Press: Washington, DC, USA, 2009; pp. 131–142. [Google Scholar] [CrossRef] [Green Version]
- Momany, M.; Taylor, I. Landmarks in the early duplication cycles of Aspergillus fumigatus and Aspergillus nidulans: Polarity, germ tube emergence and septation. Microbiology 2000, 12, 3279–3284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dörter, I.; Momany, M. Fungal Cell Cycle: A Unicellular versus Multicellular Comparison. In The Fungal Kingdom; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Hernández-Rodríguez, Y.; Momany, M. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Curr. Opin. Microbiol. 2012, 15, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.; Cowden, S.; Hernández-Rodríguez, Y.; Momany, M. Septins AspA and AspC are important for normal development and limit the emergence of new growth foci in the multicellular fungus Aspergillus nidulans. Eukaryot. Cell 2010, 9, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Vinck, A.; de Bekker, C.; Ossin, A.; Ohm, R.A.; de Vries, R.P.; Wösten, H.A.B. Heterogenic expression of genes encoding secreted proteins at the periphery of Aspergillus niger colonies. Environ. Microbiol. 2011, 13, 216–225. [Google Scholar] [CrossRef]
- Ferreira, G.F.; Santos, D.A. Heteroresistance and fungi. Mycoses 2017, 60, 562–568. [Google Scholar] [CrossRef]
- Bleichrodt, R.-J.; Foster, P.; Howell, G.; Latgé, J.-P.; Read, N.D. Cell Wall Composition Heterogeneity between Single Cells in Aspergillus fumigatus Leads to Heterogeneous Behavior during Antifungal Treatment and Phagocytosis. Mbio 2020, 11. [Google Scholar] [CrossRef]
- Harris, S.D. Morphogenesis is coordinated with nuclear division in germinating Aspergillus nidulans conidiospores. Microbiology 1999, 145 Pt 10, 2747–2756. [Google Scholar] [CrossRef] [Green Version]
- da Silva Ferreira, M.E.; Kress, M.R.V.Z.; Savoldi, M.; Goldman, M.H.S.; Härtl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Bertuzzi, M.; van Rhijn, N.; Krappmann, S.; Bowyer, P.; Bromley, M.J.; Bignell, E.M. On the lineage of Aspergillus fumigatus isolates in common laboratory use. Med. Mycol. 2020. [Google Scholar] [CrossRef]
- Latgé, J.-P.; Beauvais, A.; Chamilos, G. The Cell Wall of the Human Fungal Pathogen Aspergillus fumigatus: Biosynthesis, Organization, Immune Response, and Virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, I.; Dupres, V.; Michel, J.-P.; Duchateau, M.; Matondo, M.; Chamilos, G.; Saveanu, C.; Guijarro, J.I.; Aimanianda, V.; Lafont, F.; et al. The puzzling construction of the conidial outer layer of Aspergillus fumigatus. Cell Microbiol. 2019, 21, e12994. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Li, J.; Danion, F.; Alcazar-Fuoli, L.; Mellado, E.; Beau, R.; Jouvion, G.; Latgé, J.-P.; Fontaine, T. Two KTR Mannosyltransferases Are Responsible for the Biosynthesis of Cell Wall Mannans and Control Polarized Growth in Aspergillus fumigatus. Mbio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, T.; van Rhijn, N.; Fraczek, M.; Gsaller, F.; Davies, E.; Carr, P.; Gago, S.; Fortune-Grant, R.; Rahman, S.; Gilsenan, J.M.; et al. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat. Commun. 2020, 11, 427. [Google Scholar] [CrossRef] [Green Version]
- Muszkieta, L.; Aimanianda, V.; Mellado, E.; Gribaldo, S.; Alcàzar-Fuoli, L.; Szewczyk, E.; Prevost, M.-C.; Latgé, J.-P. Deciphering the role of the chitin synthase families 1 and 2 in the in vivo and in vitro growth of Aspergillus fumigatus by multiple gene targeting deletion. Cell Microbiol. 2014, 16, 1784–1805. [Google Scholar] [CrossRef] [PubMed]
- Ikeh, M.; Ahmed, Y.; Quinn, J. Phosphate Acquisition and Virulence in Human Fungal Pathogens. Microorganisms 2017, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, T.J.H.; Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Molecular Mechanisms of Conidial Germination in Aspergillus spp. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef]
- Ginocchio, C.C. Role of NCCLS in antimicrobial susceptibility testing and monitoring. Am. J. Health Syst. Pharm. 2002, 59, S7–S11. [Google Scholar] [CrossRef]
- Dufour, A.C.; Jonker, A.H.; Olivo-Marin, J.-C. Deciphering tissue morphodynamics using bioimage informatics. Philos. Trans. R. Soc. Lon, B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [Green Version]
- de Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Ducroz, C.; Olivo-Marin, J.-C.; Dufour, A. Spherical Harmonics based extraction and annotation of cell shape in 3D time-lapse microscopy sequences. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2011, 2011, 6619–6622. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-F.; Zhang, D.-F.; Zhang, S.; Sun, L.; Liu, Y.; Dai, J.-J. Optimizing treatment of DNA methyltransferase inhibitor RG108 on porcine fibroblasts for somatic cell nuclear transfer. Reprod. Domest. Anim. 2019, 54, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- Gosmini, R.; Nguyen, V.L.; Toum, J.; Simon, C.; Brusq, J.-M.G.; Krysa, G.; Mirguet, O.; Riou-Eymard, A.M.; Boursier, E.V.; Trottet, L.; et al. The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J. Med. Chem. 2014, 57, 8111–8131. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Saitoh, H.; Inoue, A.; Kobayashi, M.; Okitsu, Y.; Katsuoka, Y.; Fukuhara, N.; Onishi, Y.; Ishizawa, K.; Ichinohasama, R.; et al. 3-Deazaneplanocin A (DZNep), an inhibitor of S-adenosylmethionine-dependent methyltransferase, promotes erythroid differentiation. J. Biol. Chem. 2014, 289, 8121–8134. [Google Scholar] [CrossRef] [Green Version]
- Conforti, F.; Davies, E.R.; Calderwood, C.J.; Thatcher, T.H.; Jones, M.G.; Smart, D.E.; Mahajan, S.; Alzetani, A.; Havelock, T.; Maher, T.M.; et al. The histone deacetylase inhibitor, romidepsin, as a potential treatment for pulmonary fibrosis. Oncotarget 2017, 8, 48737–48754. [Google Scholar] [CrossRef]
- Kuck, D.; Caulfield, T.; Lyko, F.; Medina-Franco, J.L. Nanaomycin A selectively inhibits DNMT3B and reactivates silenced tumor suppressor genes in human cancer cells. Mol. Cancer Ther. 2010, 9, 3015–3023. [Google Scholar] [CrossRef] [Green Version]
- Takemura, Y.; Satoh, M.; Hatanaka, K.; Kubota, S. Zebularine exerts its antiproliferative activity through S phase delay and cell death in human malignant mesothelioma cells. Biosci. Biotechnol. Biochem. 2018, 82, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Wen, N.; Guo, B.; Zheng, H.; Xu, L.; Liang, H.; Wang, Q.; Wang, D.; Chen, X.; Zhang, S.; Li, Y.; et al. Bromodomain inhibitor jq1 induces cell cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. Int. J. Oncol. 2019, 55, 879–895. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiao, W.; Chen, W.; Luo, L.; Ye, S.; Liu, Y. The epigenetic modifier trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial–mesenchymal transition of lens epithelial cells. Cell Death Dis. 2013, 4, e884. [Google Scholar] [CrossRef] [Green Version]
- Lamoth, F.; Juvvadi, P.R.; Steinbach, W.J. Histone deacetylase inhibition as an alternative strategy against invasive asper-gillosis. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Wong, C.C.; Fu, L.; Chen, H.; Zhao, L.; Li, C.; Zhou, Y.; Zhang, Y.; Xu, W.; Yang, Y.; et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Sci. Transl. Med. 2018, 10, eaap9840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, D.H.; Brakhage, A.A.; Mukherjee, P.K. Gliotoxin—Bane or boon? Environ. Microbiol. 2016, 18, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Horikoshi, R.; Nakamura, S.; Mitomi, M.; Oyama, K.; Hirose, T.; Sunazuka, T.; Ōmura, S. Synthesis of pyripyropene derivatives and their pest-control efficacy. J. Pestic. Sci. 2019, 44, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, W.-R. Fumitremorgins and Relatives—From Tremorgenic Compounds to Valuable Anti-Cancer Drugs. Curr. Med. Chem. 2018, 25, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-J.; Zhang, Q.; Zhang, A.-L.; Gao, J.-M. Metabolites fromAspergillus fumigatus, an Endophytic Fungus Associated withMelia azedarach, and Their Antifungal, Antifeedant, and Toxic Activities. J. Agric. Food Chem. 2012, 60, 3424–3431. [Google Scholar] [CrossRef]
- Frisvad, J.; Larsen, T.O. Chemodiversity in the genus Aspergillus. Appl. Microbiol. Biotechnol. 2015, 99, 7859–7877. [Google Scholar] [CrossRef]
- Hobot, J.A.; Gull, K. The identification of a self-inhibitor from Syncephalastrum racemosum and its effect upon sporan-giospore germination. Antonie Van Leeuwenhoek 1980, 46, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Chitarra, G.S.; Abee, T.; Rombouts, F.M.; Posthumus, M.A.; Dijksterhuis, J. Germination of Penicillium paneum Conidia Is Regulated by 1-Octen-3-ol, a Volatile Self-Inhibitor. Appl. Environ. Microbiol. 2004, 70, 2823–2829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, N.; Moya-Nilges, M.; Sachse, M.; Locker, J.K.; Latgé, J.; Mouyna, I. Aspergillus fumigatus exoβ(1-3)glucanases family GH55 are essential for conidial cell wall morphogenesis. Cell. Microbiol. 2019, 21, e13102. [Google Scholar] [CrossRef] [PubMed]
- Novodvorska, M.; Stratford, M.; Blythe, M.J.; Wilson, R.; Beniston, R.G.; Archer, D.B. Metabolic activity in dormant conidia of Aspergillus niger and developmental changes during conidial outgrowth. Fungal Genet. Biol. 2016, 94, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, T.J.; Coolen, J.P.; Zoll, J.; Verweij, P.E.; Melchers, W. Gene co-expression analysis identifies gene clusters associated with isotropic and polarized growth in Aspergillus fumigatus conidia. Fungal Genet. Biol. 2018, 116, 62–72. [Google Scholar] [CrossRef]
- Takahashi, H.; Kusuya, Y.; Hagiwara, D.; Takahashi-Nakaguchi, A.; Sakai, K.; Gonoi, T. Global gene expression reveals stress-responsive genes in Aspergillus fumigatus mycelia. BMC Genom. 2017, 18, 1–15. [Google Scholar] [CrossRef]
- Glotzer, M. Cytokinesis in Metazoa and Fungi. Cold Spring Harb. Perspect. Biol. 2017, 9, a022343. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.E.; Gladfelter, A.S. Nuclear autonomy in multinucleate fungi. Curr. Opin. Microbiol. 2015, 28, 60–65. [Google Scholar] [CrossRef] [Green Version]
- Minke, P.F.; Lee, I.H.; Tinsley, J.H.; Bruno, K.S.; Plamann, M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 1999, 32, 1065–1076. [Google Scholar] [CrossRef]
- Nayak, T.; Edgerton-Morgan, H.; Horio, T.; Xiong, Y.; De Souza, C.P.; Osmani, S.A.; Oakley, B.R. Gamma-tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J. Cell Biol. 2010, 190, 317–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgerton-Morgan, H.; Oakley, B.R. γ-Tubulin plays a key role in inactivating APC/C(Cdh1) at the G(1)-S boundary. J. Cell Biol. 2012, 198, 785–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschuler, S.J.; Wu, L.F. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avery, S.V. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Genet. 2006, 4, 577–587. [Google Scholar] [CrossRef]
- Mela, A.P.; Momany, M. Internuclear diffusion of histone H1 within cellular compartments of Aspergillus nidulans. PLoS ONE 2018, 13, e0201828. [Google Scholar] [CrossRef]
- Kiviet, D.J.; Nghe, P.; Walker, N.; Boulineau, S.; Sunderlikova, V.; Tans, S.J. Stochasticity of metabolism and growth at the single-cell level. Nat. Cell Biol. 2014, 514, 376–379. [Google Scholar] [CrossRef]
- Hortsch, S.K.; Kremling, A. Characterization of noise in multistable genetic circuits reveals ways to modulate hetero-geneity. PLoS ONE 2018, 13, e0194779. [Google Scholar] [CrossRef] [Green Version]
- Keskin, S.; Devakanmalai, G.S.; Kwon, S.B.; Vu, H.T.; Hong, Q.; Lee, Y.Y.; Soltani, M.; Singh, A.; Ay, A.; Özbudak, E.M. Noise in the Vertebrate Segmentation Clock Is Boosted by Time Delays but Tamed by Notch Signaling. Cell Rep. 2018, 23, 2175–2185.e4. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Eser, U.; Korndorf, T.; Borsuk, M.E.; Skotheim, J.M.; Gladfelter, A.S. Nuclear repulsion enables division au-tonomy in a single cytoplasm. Curr. Biol. 2013, 23, 1999–2010. [Google Scholar] [CrossRef] [Green Version]
- Dundon, S.E.R.; Chang, S.-S.; Kumar, A.; Occhipinti, P.; Shroff, H.; Roper, M.; Gladfelter, A.S. Clustered nuclei maintain auton-omy and nucleocytoplasmic ratio control in a syncytium. Mol. Biol. Cell 2016, 27, 2000–2007. [Google Scholar] [CrossRef] [Green Version]
- Gehrmann, T.; Pelkmans, J.F.; Ohm, R.A.; Vos, A.M.; Sonnenberg, A.S.M.; Baars, J.J.P.; Wösten, H.A.B.; Reinders, M.J.T.; Abeel, T. Nu-cleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regu-latory programs. Proc. Natl. Acad. Sci. USA 2018, 115, 4429–4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, J.C.J.; Kissinger, J.C.; et al. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Bekker, C.; Bruning, O.; Jonker, M.J.; Breit, T.M.; Wösten, H.A.B. Single cell transcriptomics of neighboring hyphae of As-pergillus niger. Genome. Biol. 2011, 12, R71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mietton, F.; Ferri, E.; Champleboux, M.; Zala, N.; Maubon, D.; Zhou, Y.; Harbut, M.; Spittler, D.; Garnaud, C.; Courçon, M.; et al. Selective BET bromodomain inhibi-tion as an antifungal therapeutic strategy. Nat. Commun. 2017, 8, 15482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, A.S.; Williams, C.R.; Royce, D.B.; Pioli, P.A.; Sporn, M.B.; Liby, K.T. Bromodomain inhibitors, JQ1 and I-BET 762, as poten-tial therapies for pancreatic cancer. Cancer Lett. 2017, 394, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014, 11, 1019–1030. [Google Scholar]
- Nott, T.J.; Craggs, T.D.; Baldwin, A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular fil-ters. Nat. Chem. 2016, 8, 569–575. [Google Scholar] [CrossRef]
- Jain, S.; Parker, R. The Discovery and Analysis of P Bodies. Adv. Exp. Med. Biol. 2013, 768, 23–43. [Google Scholar] [CrossRef]
- Soukup, A.A.; Fischer, G.J.; Luo, J.; Keller, N.P. The Aspergillus nidulans Pbp1 homolog is required for normal sexual de-velopment and secondary metabolism. Fungal. Genet. Biol. 2017, 100, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Becht, P.; König, J.; Feldbrügge, M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shut-tles along microtubules. J. Cell Sci. 2006, 119, 4964–4973. [Google Scholar] [CrossRef] [Green Version]
- Morozov, I.Y.; Jones, M.G.; Spiller, D.G.; Rigden, D.J.; Dattenböck, C.; Novotny, R.; Strauss, J.; Caddick, M.X. Distinct roles for Caf1, Ccr4, Edc3 and CutA in the co-ordination of transcript deadenylation, decapping and P-body formation in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, D.; Sakai, K.; Suzuki, S.; Umemura, M.; Nogawa, T.; Kato, N.; Osada, H.; Watanabe, A.; Kawamoto, S.; Gonoi, T.; et al. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLoS ONE 2017, 12, e0177050. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, H.; Morozumi, S.; Smerage, G.H.; Teixeira, A.A. Comparison of Capillary and Test Tube Procedures for Analysis of Thermal Inactivation Kinetics of Mold Spores. J. Food Prot. 2000, 63, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Van Long, N.; Vasseur, V.; Coroller, L.; Dantigny, P.; Le Panse, S.; Weill, A.; Mounier, J.; Rigalma, K. Temperature, water activity and pH during conidia production affect the physiological state and germination time of Penicillium species. Int. J. Food Microbiol. 2017, 241, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Teertstra, W.R.; Tegelaar, M.; Dijksterhuis, J.; Golovina, E.A.; Ohm, R.A.; Wösten, H.A.B. Maturation of conidia on conidio-phores of Aspergillus niger. Fungal. Genet. Biol. 2017, 98, 61–70. [Google Scholar] [CrossRef]
- Eng, R.H.; Padberg, F.T.; Smith, S.M.; Tan, E.N.; Cherubin, C.E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef] [Green Version]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by mac-rophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef]
- Putrinš, M.; Kogermann, K.; Lukk, E.; Lippus, M.; Varik, V.; Tenson, T. Phenotypic Heterogeneity Enables Uropathogenic Escherichia coli To Evade Killing by Antibiotics and Serum Complement. Infect. Immun. 2015, 83, 1056–1067. [Google Scholar] [CrossRef] [Green Version]













| G | GA | GAP | MM | |
|---|---|---|---|---|
| Conidia area at T0, mean, px | 263 ± 9 a | 272 ± 13 a | 253 ± 21 a | 254 ± 17 a |
| Conidia area at 6 h, px | 355 ± 10 b | 378 ± 13 b | 416 ± 21 a | 519 ± 17 a |
| Conidia area at 8 h, px | 395 ± 10 c | 412 ± 13 c | 518 ± 13 b | 757 ± 21 a |
| Conidia area at 16 h, px | 471 ± 10 c | 460 ± 16 b | 1078 ± 24 a | NA |
| Conidia area at first germ tubes appearance, px | 620 ± 52 c | 545 ± 29 c | 1435 ± 39 a | 1184 ± 16 b |
| Time at first germ tube appearance, min | 1223 ± 51 a | 834 ± 28 b | 843 ± 38 b | 528 ± 15 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danion, F.; van Rhijn, N.; Dufour, A.C.; Legendre, R.; Sismeiro, O.; Varet, H.; Olivo-Marin, J.-C.; Mouyna, I.; Chamilos, G.; Bromley, M.; et al. Aspergillus fumigatus, One Uninucleate Species with Disparate Offspring. J. Fungi 2021, 7, 30. https://doi.org/10.3390/jof7010030
Danion F, van Rhijn N, Dufour AC, Legendre R, Sismeiro O, Varet H, Olivo-Marin J-C, Mouyna I, Chamilos G, Bromley M, et al. Aspergillus fumigatus, One Uninucleate Species with Disparate Offspring. Journal of Fungi. 2021; 7(1):30. https://doi.org/10.3390/jof7010030
Chicago/Turabian StyleDanion, François, Norman van Rhijn, Alexandre C. Dufour, Rachel Legendre, Odile Sismeiro, Hugo Varet, Jean-Christophe Olivo-Marin, Isabelle Mouyna, Georgios Chamilos, Michael Bromley, and et al. 2021. "Aspergillus fumigatus, One Uninucleate Species with Disparate Offspring" Journal of Fungi 7, no. 1: 30. https://doi.org/10.3390/jof7010030