In Drosophila Hemolymph, Serine Proteases Are the Major Gelatinases and Caseinases
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Biological Material
2.3. Larva Tissue Collection
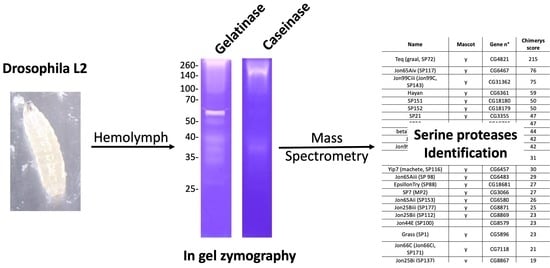
2.4. In Gel Zymography
2.5. Gelatinase Purification
2.6. Mass Spectrometry
2.7. Tequila Expression
2.8. Statistical Analysis
3. Results
3.1. Gelatinases in Drosophila melanogaster Larval Hemolymph
3.2. Larval Hemolymph Gelatinases in Different Drosophila Species
3.3. D. melanogaster Hemolymph Gelatinase and Caseinase Activities Are Inhibited by Serine Protease Inhibitors
3.4. Identification of D. melanogaster Hemolymph Putative Gelatinases and Caseinases
3.5. Hemolymph Gelatinase Activities in D. melanogaster Tequila KO Larvae
3.6. LPS Injection Did Not Affect Gelatinase Activity
3.7. Apparent Lack of Wasp Parasitism Effect on Gelatinases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wyatt, G.R. The Biochemistry of Insect Hemolymph. Annu. Rev. Entomol. 1961, 6, 75–102. [Google Scholar] [CrossRef]
- Mullins, D.E. Chemistry and Physiology of the Hemolymph. In Comprehensive Insect Physiology, Biochemistry and Pharmacology; Kerkut, G.A., Gilbert, L.I., Eds.; Integument, Respiration and Circulation; Pergamon Press: Oxford, UK, 1985; Volume 3. [Google Scholar]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, U.; Girard, J.R.; Goins, L.M.; Spratford, C.M. Drosophila as a Genetic Model for Hematopoiesis. Genetics 2019, 211, 367–417. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A.; Medzhitov, R. Innate Immune Recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tettamanti, G.; Bassal, T.; Heryanto, C.; Eleftherianos, I.; Mohamed, A. Regulators and Signalling in Insect Antimicrobial Innate Immunity: Functional Molecules and Cellular Pathways. Cell Signal. 2021, 83, 110003. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hultmark, D. Tissue Communication in a Systemic Immune Response of Drosophila. Fly 2016, 10, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.; Poirié, M.; Carton, Y. Chapter 4 The Role of Melanization and Cytotoxic By-Products in the Cellular Immune Responses of Drosophila against Parasitic Wasps. Adv. Parasit. 2009, 70, 99–121. [Google Scholar] [CrossRef]
- Dudzic, J.P.; Hanson, M.A.; Iatsenko, I.; Kondo, S.; Lemaitre, B. More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila. Cell Reports 2019, 27, 1050–1061.e3. [Google Scholar] [CrossRef] [PubMed]
- Lolo, F.-N.; Casas-Tintó, S.; Moreno, E. Cell Competition Time Line: Winners Kill Losers, Which Are Extruded and Engulfed by Hemocytes. Cell Reports 2012, 2, 526–539. [Google Scholar] [CrossRef]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Klein, T.; Bischoff, R. Physiology and Pathophysiology of Matrix Metalloproteases. Amino Acids 2011, 41, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; den Steen, P.E.V.; Opdenakker, G. Biochemistry and Molecular Biology of Gelatinase B or Matrix Metalloproteinase-9 (MMP-9): The next Decade. Crit. Rev. Biochem. Mol. 2013, 48, 222–272. [Google Scholar] [CrossRef]
- Descamps, F.J.; Martens, E.; Opdenakker, G. Analysis of Gelatinases in Complex Biological Fluids and Tissue Extracts. Lab. Investig. 2002, 82, 1607–1608. [Google Scholar] [CrossRef] [PubMed]
- Ligi, D.; Maniscalco, R.; Mannello, F. MMP-2 and MMP-9 in Human Peripheral Blood: Optimizing Gelatinase Calibrator for Degradome Research and Discovering a Novel Gelatinolytic Enzyme. J. Proteome Res. 2020, 19, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Saito, K.; Kunisaki, N.; Kimura, S. Green Tea Polyphenols Inhibit Metalloproteinase Activities in the Skin, Muscle, and Blood of Rainbow Trout. J. Agric. Food Chem. 2002, 50, 7169–7174. [Google Scholar] [CrossRef] [PubMed]
- Mannello, F.; Canesi, L.; Gazzanelli, G.; Gallo, G. Biochemical Properties of Metalloproteinases from the Hemolymph of the Mussel Mytilus Galloprovincialis Lam. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 128, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Llano, E.; Adam, G.; Pendás, A.M.; Quesada, V.; Sánchez, L.M.; Santamaría, I.; Noselli, S.; López-Otín, C. Structural and Enzymatic Characterization of Drosophila Dm2-MMP, a Membrane-Bound Matrix Metalloproteinase with Tissue-Specific Expression. J. Biol. Chem. 2002, 277, 23321–23329. [Google Scholar] [CrossRef] [PubMed]
- LaFever, K.S.; Wang, X.; Page-McCaw, P.; Bhave, G.; Page-McCaw, A. Both Drosophila Matrix Metalloproteinases Have Released and Membrane-Tethered Forms but Have Different Substrates. Sci. Rep. 2017, 7, 44560. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L.J.; Page-McCaw, A. A Secreted MMP Is Required for Reepithelialization during Wound Healing. Mol. Biol. Cell. 2012, 23, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Chen, Z.; Zhang, Z.; Jia, Q. The Expression, Purification, and Substrate Analysis of Matrix Metalloproteinases in Drosophila melanogaster. Protein Expr. Purif. 2020, 171, 105629. [Google Scholar] [CrossRef]
- Woodhouse, E.; Hersperger, E.; Stetler-Stevenson, W.G.; Liotta, L.A.; Shearn, A. Increased Type IV Collagenase in Lgl-Induced Invasive Tumors of Drosophila. Cell Growth Differ. 1994, 5, 151–159. [Google Scholar] [PubMed]
- Bylemans, D.; Paemen, L.; Huybrechts, R.; Odenakker, G.; Loof, A.D. Sex, and Developmental Stage-Specific Gelatinolytic Activity in the Fleshfly Neobellieria bullata and the Regulating Role of 20-OH-Ecdysone. Comp. Biochem. Physiol. 1997, 118A, 1327–1333. [Google Scholar] [CrossRef]
- Dubuffet, A.; Colinet, D.; Anselme, C.; Dupas, S.; Carton, Y.; Poirié, M. Variation of Leptopilina boulardi Success in Drosophila Hosts What is Inside the Black Box? Chapter 6. Adv. Parasitol. 2009, 70, 147–188. [Google Scholar] [CrossRef] [PubMed]
- Munier, A.I.; Medzhitov, R.; Janeway, C.A.; Doucet, D.; Capovilla, M.; Lagueux, M. Graal: A Drosophila Gene Coding for Several Mosaic Serine Proteases. Insect Biochem. Mol. Biol. 2004, 34, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Chabert, S.; Allemand, R.; Poyet, M.; Eslin, P.; Gibert, P. Ability of European Parasitoids (Hymenoptera) to Control a New Invasive Asiatic Pest, Drosophila suzukii. Biol. Control 2012, 63, 40–47. [Google Scholar] [CrossRef]
- Wan, B.; Goguet, E.; Ravallec, M.; Pierre, O.; Lemauf, S.; Volkoff, A.-N.; Gatti, J.-L.; Poirié, M. Venom Atypical Extracellular Vesicles as Interspecies Vehicles of Virulence Factors Involved in Host Specificity: The Case of a Drosophila Parasitoid Wasp. Front. Immunol. 2019, 10, 1688. [Google Scholar] [CrossRef] [PubMed]
- Métayer, S.; Dacheux, F.; Dacheux, J.-L.; Gatti, J.-L. Comparison, Characterization, and Identification of Proteases and Protease Inhibitors in Epididymal Fluids of Domestic Mammals. Matrix Metalloproteinases Are Major Fluid Gelatinases. Biol. Reprod. 2002, 66, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.H. Silver Stain for Proteins in Polyacrylamide Gels: A Modified Procedure with Enhanced Uniform Sensitivity. Anal. Biochem. 1981, 117, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.B.; Wolfe, J.; Akam, M.E. The Developmental Profiles of Two Major Haemolymph Proteins from Drosophila melanogaster. J. Insect Physiol. 1977, 23, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Akam, M.E.; Roberts, D.B.; Wolfe, J. Drosophila Hemolymph Proteins: Purification, Characterization, and Genetic Mapping of Larval Serum Protein 2 in D. Melanogaster. Biochem. Genet. 1978, 16, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Miguel-Aliaga, I. The Digestive Tract of Drosophila melanogaster. Annu. Rev. Genet. 2013, 47, 377–404. [Google Scholar] [CrossRef] [PubMed]
- Vierstraete, E.; Verleyen, P.; Baggerman, G.; D’Hertog, W.; den Bergh, G.V.; Arckens, L.; Loof, A.D.; Schoofs, L. A Proteomic Approach for the Analysis of Instantly Released Wound and Immune Proteins in Drosophila melanogaster Hemolymph. Proc. Natl. Acad. Sci. USA 2004, 101, 470–475. [Google Scholar] [CrossRef]
- Karlsson, C.; Korayem, A.M.; Scherfer, C.; Loseva, O.; Dushay, M.S.; Theopold, U. Proteomic Analysis of the Drosophila Larval Hemolymph Clot. J. Biol. Chem. 2004, 279, 52033–52041. [Google Scholar] [CrossRef] [PubMed]
- Levy, F.; Bulet, P.; Ehret-Sabatier, L. Proteomic Analysis of the Systemic Immune Response of Drosophila. Molecular & cellular proteomics. Mol. Cell Proteom. 2004, 3, 156–166. [Google Scholar] [CrossRef]
- Imler, J.-L.; Tauszig, S.; Jouanguy, E.; Forestier, C.; Hoffmann, J.A. LPS-Induced Immune Response in Drosophila. J. Endotoxin Res. 2000, 6, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Istas, O.; Greenhalgh, A.; Cooper, R. The Effects of a Bacterial Endotoxin on Behavior and Sensory-CNS-Motor Circuits in Drosophila melanogaster. Insects 2019, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Carton, Y.; Bouletreau, M.; Alphen, J.; van Lenteren, J. The Drosophila Parasitic Wasps. In The Genetics and Biology of Drosophila; Ashburner, M., Carson, J.H.L., Thompson, J.N., Eds.; Academic Press: London, UK, 1986; Volume 3e, pp. 347–394. [Google Scholar]
- Colinet, D.; Dubuffet, A.; Cazes, D.; Moreau, S.; Drezen, J.-M.; Poirié, M. A Serpin from the Parasitoid Wasp Leptopilina Boulardi Targets the Drosophila Phenoloxidase Cascade. Dev. Comp. Immunol. 2009, 33, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R.; Jiang, H. Clip-Domain Serine Proteases as Immune Factors in Insect Hemolymph. Curr. Opin. Insect Sci. 2015, 11, 47–55. [Google Scholar] [CrossRef]
- Kambris, Z.; Brun, S.; Jang, I.-H.; Nam, H.-J.; Romeo, Y.; Takahashi, K.; Lee, W.-J.; Ueda, R.; Lemaitre, B. Drosophila Immunity: A Large-Scale In Vivo RNAi Screen Identifies Five Serine Proteases Required for Toll Activation. Curr. Biol. 2006, 16, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Poidevin, M.; Kwon, H.-M.; Guillou, A.; Sottas, V.; Lee, B.L.; Lemaitre, B. A Single Modular Serine Protease Integrates Signals from Pattern-Recognition Receptors Upstream of the Drosophila Toll Pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 12442–12447. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Hutter, S.; Stamboliyska, R.; Saminadin-Peter, S.S.; Stephan, W.; Parsch, J. Population transcriptomics of Drosophila melanogaster females. BMC Genom. 2011, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.R.; Dziedziech, A.; Arefin, B.; Kienzle, T.; Wang, Z.; Akhter, M.; Berka, J.; Theopold, U. Insect Hemolymph Coagulation: Kinetics of Classically and Non-Classically Secreted Clotting Factors. Insect Biochem. Mol. Biol. 2019, 109, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jiang, H. Building a Platform for Predicting Functions of Serine Protease-Related Proteins in Drosophila melanogaster and other Insects. Insect Biochem. Mol. Biol. 2018, 103, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Mhetre, A.; Ratnaparkhi, G.S.; Kamat, S.S. A Superfamily-Wide Activity Atlas of Serine Hydrolases in Drosophila melanogaster. Biochemistry 2021, 60, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-W.; Wang, H.-D.; Bai, H.; Wu, M.-S.; Yen, J.-H.; Tatar, M.; Fu, T.-F.; Wang, P.-Y. Tequila Regulates Insulin-Like Signaling and Extends Life Span in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.A.; Juarez, M.T.; Hermann, A.; Sasik, R.; Hardiman, G.; McGinnis, W. Serine Proteolytic Pathway Activation Reveals an Expanded Ensemble of Wound Response Genes in Drosophila. PLoS ONE 2013, 8, e61773. [Google Scholar] [CrossRef] [PubMed]
- Levy, F. Analyse de la Réponse Immunitaire de la Drosophile par une Approche Protéomique. Thèse, Université Louis Pasteur, Strasbourg, I. Faculté des Sciences de la Vie, Strasbourg, France, 2005. Available online: https://publication-theses.unistra.fr/public/theses_doctorat/2005/LEVY_Francine_2005.pdf (accessed on 30 October 2023).
- Carlson, J.R.; Hogness, D.S. Developmental and Functional Analysis of Jonah Gene Expression. Dev. Biol. 1985, 108, 355–368. [Google Scholar] [CrossRef]
- Christesen, D.; Yang, Y.T.; Somers, J.; Robin, C.; Sztal, T.; Batterham, P.; Perry, T. Transcriptome Analysis of Drosophila melanogaster Third Instar Larval Ring Glands Points to Novel Functions and Uncovers a Cytochrome P450 Required for Development. G3 Genes Genomes Genet. 2016, 7, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Lipinszki, Z.; Klement, E.; Hunyadi-Gulyas, E.; Medzihradszky, K.F.; Márkus, R.; Pál, M.; Deák, P.; Udvardy, A. A Novel Interplay between the Ubiquitin–Proteasome System and Serine Proteases during Drosophila Development. Biochem. J. 2013, 454, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Eleftherianos, I. Participation of the Serine Protease Jonah66Ci in the Drosophila Antinematode Immune Response. Infect. Immun. 2019, 87, e00094-19. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, N.; Hasegawa, S.; Nagashima, Y.; Mitsuhashi, K.; Tsubota, Y.; Miyata, S.; Miyagi, Y.; Yasumitsu, H.; Miyazaki, K. Expression of Trypsin by Epithelial Cells of Various Tissues, Leukocytes, and Neurons in Human and Mouse. Am. J. Pathol. 1998, 153, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Aliaga, I.; Jasper, H.; Lemaitre, B. Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 2018, 210, 357–396. [Google Scholar] [CrossRef] [PubMed]
- Pilpel, N.; Nezer, I.; Applebaum, S.W.; Heifetz, Y. Mating-Increases Trypsin in Female Drosophila Hemolymph. Insect Biochem. Mol. Biol. 2008, 38, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Chintapalli, V.R.; Wang, J.; Dow, J.A.T. Using FlyAtlas to Identify Better Drosophila melanogaster Models of Human Disease. Nat. Genet. 2007, 39, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.K.; Tripathi, L.P.; Jensen, L.J.; Gahnim, M.; Mason, C.; Furlong, E.E.; Rodrigues, V.; White, K.P.; Bork, P.; Sowdhamini, R. Enhanced Function Annotations for Drosophila Serine Proteases: A Case Study for Systematic Annotation of Multi-Member Gene Families. Gene 2008, 407, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Gettins, P.G.W. Serpin Structure, Mechanism, and Function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef] [PubMed]
- Romejko, K.; Markowska, M.; Niemczyk, S. The Review of Current Knowledge on Neutrophil Gelatinase-Associated Lipocalin (NGAL). Int. J. Mol. Sci. 2023, 24, 10470. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.R.; Yerbury, J.J.; Ecroyd, H.; Wilson, M.R. Extracellular Chaperones and Proteostasis. Biochemistry 2013, 82, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Serra, N.D.; Sundaram, M.V. Transcytosis in the Development and Morphogenesis of Epithelial Tissues. EMBO J. 2021, 40, e106163. [Google Scholar] [CrossRef] [PubMed]





| D. melanogaster | D. immigrans | D. yakuba | D. simulans | D. suzukii | ||
|---|---|---|---|---|---|---|
| Nasrallah | Canton S | |||||
| E64 | −−− | −−− | −−− | −−− | −−− | −−− |
| AEBSF | +++ | −++ | +++ | +++ | +++ | +++ |
| Leupeptin | +++ | +++ | −++ | −++ | +++ | +++ |
| EDTA | −−− | −−− | −−− | −−− | −−− | −−− |
| Name | CG | Mascot Score | Type | |
|---|---|---|---|---|
| Band G1 | SP117/Jon65Aiv | CG6467 | 396 | Chymotrypsin-like serine protease |
| SP98/Jon65Aiii | CG6483 | 245 | Elastase-like serine protease | |
| SP72/Tequila/Graal | CG4821 | 216 | Trypsin-like serine protease | |
| SP21 | CG3355 | 118 | Trypsin-like serine protease | |
| SP131 | CG17475 | 65 | Chymotrypsin-like serine protease | |
| SP172/modSP | CG31217 | 63 | Chymotrypsin-like serine protease | |
| SP119/betaTry | CG18211 | 56 | Trypsin-like serine protease | |
| Hayan | CG6361 | 55 | Trypsin-like serine protease | |
| Band G2 | SP72/Tequila/Graal | CG4821 | 744 | Trypsin-like serine protease |
| SP98/Jon65Aiii | CG6483 | 461 | Elastase-like serine protease | |
| Hayan | CG6361 | 256 | Trypsin-like serine protease | |
| SP117/Jon65Aiv | CG6467 | 211 | Chymotrypsin-like serine protease | |
| SP172/modSP | CG31217 | 148 | Chymotrypsin-like serine protease | |
| SP21 | CG3355 | 137 | Trypsin-like serine protease | |
| SP91 | CG16749 | 103 | Chymotrypsin-like serine protease | |
| SP131 | CG17475 | 76 | Chymotrypsin-like serine protease | |
| SP119/betaTry | CG18211 | 71 | Trypsin-like serine protease | |
| SP222 | CG30187 | 70 | Trypsin-like serine protease | |
| SP88/epsilonTry | CG18681 | 69 | Trypsin-like serine protease | |
| SP143/Jon99Ciii | CG31362 | 54 | Chymotrypsin-like serine protease | |
| Band G3 | SP72/Tequila/Graal | CG4821 | 816 | Trypsin-like serine protease |
| SP98/Jon65Aiii | CG6483 | 460 | Elastase-like serine protease | |
| SP143/Jon99Ciii | CG31362 | 248 | Chymotrypsin-like serine protease | |
| SP119/betaTry | CG18211 | 216 | Trypsin-like serine protease | |
| SP117/Jon65Aiv | CG6467 | 139 | Chymotrypsin-like serine protease | |
| SP102/alphaTry | CG18444 | 114 | Trypsin-like serine protease | |
| SP21 | CG3355 | 104 | Trypsin-like serine protease | |
| SP29 | CG14642 | 83 | Trypsin-like serine protease | |
| psh/persephone | CG6367 | 77 | Trypsin-like serine protease | |
| SP25/Melanization Protease 1 | CG1102 | 70 | Trypsin-like serine protease | |
| SP145/Jon65Ai | CG10475 | 58 | Chymotrypsin-like serine protease | |
| SP52 | CG8952 | 56 | Elastase-like serine protease | |
| Band G4 | SP151 | CG18180 | 459 | Elastase-like serine protease |
| SP98/Jon65Aiii | CG6483 | 386 | Elastase-like serine protease | |
| SP143/Jon99Ciii | CG31362 | 309 | Chymotrypsin-like serine protease | |
| SP88/epsilonTry | CG18681 | 297 | Trypsin-like serine protease | |
| SP117/Jon65Aiv | CG6467 | 295 | Chymotrypsin-like serine protease | |
| SP72/Tequila/Graal | CG4821 | 277 | Trypsin-like serine protease | |
| SP171/Jon66Ci | CG7118 | 267 | Elastase-like serine protease | |
| SP181/Jon99Fi | CG18030 | 229 | Chymotrypsin-like serine protease | |
| SP116/yip7/machete | CG6457 | 210 | Chymotrypsin-like serine protease | |
| SP177/Jon25Biii | CG8871 | 196 | Chymotrypsin-like serine protease | |
| SP152 | CG18179 | 188 | Chymotrypsin-like serine protease | |
| SP78 | CG10472 | 180 | Chymotrypsin-like serine protease | |
| SP165/Jon66Cii | CG7170 | 163 | Chymotrypsin-like serine protease | |
| SP83 | CG17571 | 152 | Trypsin-like serine protease | |
| SP119/betaTry | CG18211 | 143 | Trypsin-like serine protease | |
| SP139/thetaTry | CG12385 | 141 | Trypsin-like serine protease | |
| SP153/Jon65Aii | CG6580 | 140 | Elastase-like serine protease | |
| SP177/Jon25Bii | CG8869 | 132 | Chymotrypsin-like serine protease | |
| SP40 | CG4613 | 59 | Trypsin-like serine protease | |
| SP118 | CG34458 | 53 | Chymotrypsin-like serine protease |
| Name | CG | Mascot Score | Type | |
|---|---|---|---|---|
| Band C1 | SP117/Jon65Aiv | CG6467 | 72 | Chymotrypsin-like serine protease |
| SP72/Tequila/Graal | CG4821 | 50 | Trypsin-like serine protease | |
| Band C2 | SP117/Jon65Aiv | CG6467 | 446 | Chymotrypsin-like serine protease |
| SP21 | CG3355 | 137 | Trypsin-like serine protease | |
| Band C3 | SP72/Tequila/Graal | CG4821 | 797 | Trypsin-like serine protease |
| SP143/Jon99Ciii | CG31362 | 557 | Chymotrypsin-like serine protease | |
| SP145/Jon65Ai | CG10475 | 346 | Chymotrypsin-like serine protease | |
| SP177/Jon25Bii | CG8869 | 330 | Chymotrypsin-like serine protease | |
| SP137/Jon25Bi | CG8867 | 291 | Elastase-like serine protease | |
| c-SP1/grass | CG5896 | 254 | Trypsin-like serine protease | |
| SP181/Jon99Fi | CG18030 | 254 | Chymotrypsin-like serine protease | |
| SP116/yip7/machete | CG6457 | 208 | Chymotrypsin-like serine protease | |
| SP177/Jon25Biii | CG8871 | 174 | Chymotrypsin-like serine protease | |
| SP33/spirit | CG2056 | 173 | Trypsin-like serine protease | |
| SP34 | CG9372 | 159 | Trypsin-like serine protease | |
| SP7/Melanization Protease 2 | CG3066 | 151 | Trypsin-like serine protease | |
| SP117/Jon65Aiv | CG6467 | 132 | Chymotrypsin-like serine protease | |
| SP21 | CG3355 | 122 | Trypsin-like serine protease | |
| SP25/Melanization Protease 1 | CG1102 | 114 | Trypsin-like serine protease | |
| SP153/Jon65Aii | CG6580 | 111 | Elastase-like serine protease | |
| Hayan | CG6361 | 100 | Trypsin-like serine protease | |
| SP171/Jon66Ci | CG7118 | 69 | Elastase-like serine protease | |
| SP91 | CG16749 | 68 | Chymotrypsin-like serine protease |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatti, J.-L.; Lemauf, S.; Belghazi, M.; Arthaud, L.; Poirié, M. In Drosophila Hemolymph, Serine Proteases Are the Major Gelatinases and Caseinases. Insects 2024, 15, 234. https://doi.org/10.3390/insects15040234
Gatti J-L, Lemauf S, Belghazi M, Arthaud L, Poirié M. In Drosophila Hemolymph, Serine Proteases Are the Major Gelatinases and Caseinases. Insects. 2024; 15(4):234. https://doi.org/10.3390/insects15040234
Chicago/Turabian StyleGatti, Jean-Luc, Séverine Lemauf, Maya Belghazi, Laury Arthaud, and Marylène Poirié. 2024. "In Drosophila Hemolymph, Serine Proteases Are the Major Gelatinases and Caseinases" Insects 15, no. 4: 234. https://doi.org/10.3390/insects15040234