Mass-Rearing Conditions Do Not Always Reduce Genetic Diversity: The Case of the Mexican Fruit Fly, Anastrepha ludens (Diptera: Tephritidae)
Abstract
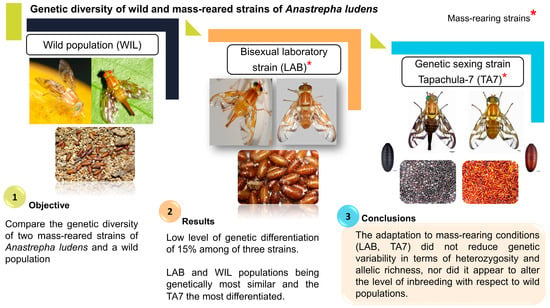
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Extraction and Amplification of Nuclear DNA
2.3. Data Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernández-Ortíz, V.; Aluja, M. Listado de especies del género Neotropical Anastrepha (Diptera: Tephritidae) con notas sobre su distribución y plantas hospederas. Folia Entomol. Mex. 1993, 88, 89–105. [Google Scholar]
- Dias, N.P.; Zotti, M.J.; Montoya, P.; Carvalho, I.R.; Nava, D.E. Fruit fly management research: A systematic review of monitoring and control tactics in the world. Crop Prot. 2018, 112, 187–200. [Google Scholar] [CrossRef]
- APHIS. Anastrepha ludens, Mexican Fruit Fly Host List. Available online: https://www.aphis.usda.gov/plant_health/plant_pest_info/fruit_flies/downloads/host-lists/mexfly-host-list.pdf (accessed on 22 February 2023).
- Reyes, J.; Santiago, G.; Hernández, P. The Mexican fruit fly eradication programme. In Area-Wide Control of Fruit Flies and Other Insect Pests; Tan, K.H., Ed.; Penerbit Universiti Sains Malaysia: Penang, Malaysia, 2000; pp. 377–380. [Google Scholar]
- Gutiérrez, J.M. El programa Moscas de la Fruta en México. In Moscas de la Fruta: Fundamentos y Procedimientos Para su Manejo; Montoya, P., Toledo, J., Hernández, E., Eds.; S y G Editores: Coyoacán, México, 2010; pp. 3–9. [Google Scholar]
- Knipling, E.F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 1955, 48, 459–462. [Google Scholar] [CrossRef]
- Orozco-Dávila, D.; Quintero, L.; Hernández, E.; Solís, E.; Artiaga, T.; Hernández, R.; Ortega, C.; Montoya, P. Mass rearing and sterile insect releases for the control of Anastrepha spp. pests in Mexico—A review. Entomol. Exp. App. 2017, 164, 176–187. [Google Scholar] [CrossRef]
- Hendrichs, J.; Franz, G.; Rendon, P. Increased effectiveness and applicability of the sterile insect technique through male-only releases for control of Mediterranean fruit flies during fruiting seasons. J. Appl. Entomol. 1995, 119, 371–377. [Google Scholar] [CrossRef]
- Caceres, C.; Cayol, J.P.; Enkerlin, W.R.; Franz, G.; Hendrichs, J.; Robinson, A.S. Comparison of Mediterranean fruit fly (Ceratitis capitata) (Tephritidae) bisexual and genetic sexing strains: Development, evaluation and economics In Proceedings, Symposium: 6th International Symposium on Fruit Flies of Economic Importance; Barnes, B.N., Ed.; Isteg Scientific Publications: Irene, South Africa, 2004; pp. 367–381. [Google Scholar]
- Orozco, D.; Hernández, M.R.; Meza, J.S.; Quintero, J.L. Do sterile females affect the sexual performance of sterile males of Anastrepha ludens (Diptera: Tephritidae)? J. Appl. Entomol. 2013, 137, 321–326. [Google Scholar] [CrossRef]
- Mclnnis, D.O.; Tarn, S.; Grace, C.; Miyashita, D. Population suppression and sterility rates induced by variable sex ratio, sterile insect releases of Ceratitis capitata (Diptera: Tephritidae) in Hawaii. Ann. Entomol. Soc. Am. 1994, 87, 231–240. [Google Scholar] [CrossRef]
- Rendón, P.; McInnis, D.; Lance, D.; Stewart, J. Comparison of Medfly male-only and bisexual releases in large scale field trials. In Area-Wide Control of Fruit Flies and Other Insect Pests; Tan, K.H., Ed.; Penerbit Universiti Sains Malaysia: Penang, Malaysia, 2000; pp. 517–525. [Google Scholar]
- Quintero-Fong, L.; Luis, J.H.; Montoya, P.; Orozco-Dávila, D. In situ sexual competition between sterile males of the genetic sexing Tapachula-7 strain and wild Anastrepha ludens flies. Crop Prot. 2018, 106, 1–5. [Google Scholar] [CrossRef]
- Orozco, D.; Meza, J.S.; Zepeda, S.; Solís, E.; Quintero-Fong, J.L. Tapachula-7, a new genetic sexing strain of the Mexican fruit fly (Diptera: Tephritidae): Sexual compatibility and competitiveness. J. Econ. Entomol. 2013, 106, 735–741. [Google Scholar] [CrossRef]
- Zepeda-Cisneros, C.S.; Meza Hernández, J.S.; García-Martínez, V.; Ibañez Palacios, J.; Zacharopoulou, A.; Franz, G. Development, genetic and cytogenetic analyses of genetic sexing strains of the Mexican fruit fly, Anastrepha ludens Loew (Diptera: Tephritidae). BMC Genet. Data 2014, 15 (Suppl. S2), S1. [Google Scholar] [CrossRef]
- Franz, G.; Gencheva, E.; Kerremans, P.H. Improved stability of genetic sex separation strains for the Mediterranean fruit fly, Ceratitis capitata. Genome 1994, 37, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.; Caceres, C. A filter rearing system for mass reared sexing strains of Mediterranean fruit fly (Diptera: Tephritidae). In Area-Wide Control of Fruit Flies and Other Pests; Tan, K.H., Ed.; Penerbit Universiti Sains Malaysia: Penang, Malaysia, 2000; pp. 543–550. [Google Scholar]
- Woodworth, L.M.; Montgomery, M.E.; Briscoe, D.A.; Frankham, R. Rapid genetic deterioration in captive populations: Causes and conservation implications. Conserv. Genet. 2002, 3, 277–288. [Google Scholar] [CrossRef]
- Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 2008, 17, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, A.S.; Cameron, E.C.; Sved, J.A.; Meats, A.W. Genetic consequences of domestication and mass rearing of pest fruit fly Bactrocera tryoni (Diptera: Tephritidae). J. Econ. Entomol. 2012, 105, 1051–1056. [Google Scholar] [CrossRef]
- Meza, J.S.; Arredondo, J.; Orozco, D.; Pérez-Staples, D. Disparity in sexual behaviour between wild and mass-reared Mexican fruit flies. Physiol. Entomol. 2014, 39, 263–270. [Google Scholar] [CrossRef]
- Parreño, M.A.; Scannapieco, A.C.; Remis, M.I.; Juri, M.; Vera, M.T.; Segura, D.F.; Cladera, J.L.; Lanzavecchia, S.B. Dynamics of genetic variability in Anastrepha fraterculus (Diptera: Tephritidae) during adaptation to laboratory rearing conditions. BMC Genet. Data 2014, 15, S14. [Google Scholar] [CrossRef] [PubMed]
- Krafsur, E.S.; Ouma, J.O. Role of population genetics in the sterile insect technique. In Sterile insect Technique: Principles and Practice in Area-Wide Integrated Pest Management, 2nd ed.; Dick, V.A., Hendrichs, J., Robinson, A.S., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 529–548. [Google Scholar]
- Romiguier, J.; Gayral, P.; Ballenghien, M.; Bernard, A.; Cahais, V.; Chenuil, A.; Chiari, Y.; Dernat, R.; Duret, L.; Faivre, N.; et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 2014, 515, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Bush, G.L.; Neck, R.W.; Kitto, B.G. Screwworm eradicaction: Inadvertent selection for noncompetitive ecotypes during mass rearing. Science 1976, 193, 492–493. [Google Scholar] [CrossRef]
- Loukas, M.; Economopoulos, A.P.; Zouros, E.; Vergini, Y. Genetic changes in artificially reared colonies of the olive fruit fly (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 1985, 78, 159–165. [Google Scholar] [CrossRef]
- Simões, P.; Pascual, M.; Santos, J.; Rose, R.M.; Matos, M. Evolutionary dynamics of molecular markers during local adaptation: A case study in Drosophila subobscura. BMC Evol. Biol. 2008, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Tsakas, S.C.; Zouros, E. Genetic differences among natural and laboratory reared populations of the olive-fruit fly Dacus oleae (Diptera: Tephritidae). Entomol. Exp. Appl. 1980, 28, 268–276. [Google Scholar] [CrossRef]
- Economopoulos, A.P.; Loukas, M. ADH allele frequency changes in olive fruit flies shift from olives to artificial larval food and vice versa, effect of temperature. Entomol. Exp. Appl. 1986, 40, 215–221. [Google Scholar] [CrossRef]
- Zygouridis, N.E.; Argov, Y.; Nemny-Lavy, E.; Augustinos, A.A.; Nestel, D.; Mathiopoulos, K.D. Genetic changes during laboratory domestication of an olive fly SIT strain. J. Appl. Entomol. 2014, 138, 423–432. [Google Scholar] [CrossRef]
- Orozco-Dávila, D.; Hernández, R.; Meza, S.; Domínguez, J. Sexual competitiveness and compatibility between mass-reared sterile flies and wild populations of Anastrepha ludens (Diptera: Tephritidae) from different regions in Mexico. Fla. Entomol. 2007, 90, 19–26. [Google Scholar] [CrossRef]
- Caceres, C. Mass rearing of temperature sensitive genetic sexing strains in the Mediterranean fruit fly (Ceratitis capitata). Genetica 2002, 116, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Fritz, A.H.; Schable, N. Microsatellite loci from the Caribbean Fruit Fly, Anastrepha suspensa (Diptera: Tephritidae). Mol. Ecol. Notes 2004, 4, 443–445. [Google Scholar] [CrossRef]
- Morin, P.A.; Manaster, C.; Mesnick, S.L.; Holland, R. Normalization and binning of historical and multi-source microsatellite data: Overcoming the problems of allele size shift with allelogram. Mol. Ecol. Resour. 2009, 9, 1451–1455. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetics software for teaching and research. Mol. Ecol. 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. Biometrics, 2nd ed.; Freeman and Company: New York, NY, USA, 1995; p. 794. [Google Scholar]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.M.; Estoup, A. DYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A.; Zhang, W.; Balding, D.J. Approximate Bayesian computation in population genetics. Genetics 2002, 162, 2025–2035. [Google Scholar] [CrossRef]
- Estoup, A.; Lombaert, E.; Marin, J.M.; Guillemaud, T.; Pudlo, P.; Robert, C.P.; Cornuet, J.M. Estimation of demo-genetic model probabilities with approximates Bayesian computation using linear discriminant analysis on summary statistics. Mol. Ecol. Resour. 2012, 12, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Haymer, D.S. Genetic analysis of laboratory and wild strains of the Melon fly (Diptera: Tephritidae) using random amplified polymorphic DNA–polymerase chain reaction. Ann. Entomol. Soc. Am. 1995, 88, 705–710. [Google Scholar] [CrossRef]
- Hernández, E.; Toledo, J.; Artiaga-López, T.; Flores, S. Demographic changes in Anastrepha obliqua (Diptera: Tephritidae) throughout the laboratory colonization process. J. Econ. Entomol. 2009, 102, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Bryant, E.H.; McCommas, S.A.; Combs, L.M. The effect of an experimental bottleneck upon quantitative genetic variation in the Housefly. Genetics 1986, 114, 1191–1211. [Google Scholar] [CrossRef]
- Zouros, E.; Loukas, M.; Economopoulos, A.; Mazomenos, B. Selection at the alcohol dehydrogenase locus of the olive fruit fly Dacus oleae under artificial rearing. Heredity 1982, 48, 169–185. [Google Scholar] [CrossRef]
- Molina-Nery, M.C.; Ruiz-Montoya, L.; Zepeda-Cisneros, C.S.; Liedo, P. Genetic structure of populations of Anastrepha ludens (Diptera: Tephritidae) in Mexico. Fla. Entomol. 2014, 97, 1648–1661. [Google Scholar] [CrossRef]
- Ruiz-Montoya, L.; Verónica-Vallejo, R.; Haymer, D.; Liedo, P. Genetic and ecological relationships of Anastrepha ludens (Diptera: Tephritidae) populations in souther Mexico. Insects 2020, 11, 815. [Google Scholar] [CrossRef]
- Gálvez-Reyes, N.G.; Salvador-Figueroa, M.; Santini, N.S.; Mastretta-Yanes, A.; Núñez-Farfán, J.; Piñero, D. Nuclear genetic diversity and structure of Anastrepha ludens wild populations evidenced by microsatellite markers. Front. Ecol. Evol. 2022, 10, 948640. [Google Scholar] [CrossRef]




| Markers | Primer Sequence (5′-3′) | Size (bp) | Temp (°C) |
|---|---|---|---|
| Asus1-1H | 1HF- TGG TAG TCA GGC ATC AA | 208–230 | 54 |
| 1HR- CAG TAT ATC TTG GGC AAT AA | |||
| Asus1-2B | 2BF- CAG CAG CGT ATG TAT GT | 129–132 | 54 |
| 2BR- CTT CGG TGT TGT TTA CTT A | |||
| Asus1-2F | 2FF- GCC ACT GGT TTA TTA CTC T | 262–286 | 52 |
| 2FR- ACG CCA GAC ATT TTA GTT | |||
| Asus1-3B | 3BF- CGT TCA GCA TTA CTT TGA | 107–123 | 54 |
| 3BR- CTT ATT TGG AAG TGA CTGA | |||
| Asus1-4H | 4HF- TGC CAT GTC TTG CTA GT | 158–204 | 52 |
| 4HR- TTA CCC TGA CTG ATT GTT AT | |||
| Asus1-5E | 5EF- CAA CCC GAT TCA GAT TA | 235–275 | 52 |
| 5ER- CGA AAA ATC CAA ATA TCT TA | |||
| Asus1-6C | 6CF- AAA TCG TGG TAA ATA AAG TAA C | 333–377 | 54 |
| 6CR- CGC TGC TCA ATT TAA TAC T | |||
| Asus1-8D | 8DF- GTT AAG CCA TTC CTG TTC | 215–254 | 53 |
| 8DR- CTG ACA GGG CAA AGT TAC | |||
| Asus1-9A | 9AF- AAA CCA TAC TTG AGA AAA AC | 278–329 | 49 |
| 9AR- TTG GAA CGA GAA TAA AAC |
| Strain | Locus | DF | ChiSq | p |
|---|---|---|---|---|
| LAB | ASUS1-1H | 3 | 60.000 | <0.01 |
| ASUS1-2B | 10 | 75.820 | <0.01 | |
| ASUS1-2F | 15 | 73.972 | <0.01 | |
| ASUS1-3B | 6 | 56.000 | <0.01 | |
| ASUS1-4H | 10 | 90.015 | <0.01 | |
| ASUS1-6C | 3 | 11.422 | 0.010 | |
| ASUS1-8D | 10 | 52.095 | <0.01 | |
| TA7 | ASUS1-1H | 3 | 30.267 | <0.01 |
| ASUS1-2B | 6 | 31.516 | <0.01 | |
| ASUS1-2F | 3 | 21.250 | <0.01 | |
| ASUS1-3B | 1 | 30.000 | <0.01 | |
| ASUS1-4H | 1 | 15.648 | <0.01 | |
| ASUS1-6C | 15 | 12.990 | 0.603 | |
| ASUS1-8D | 21 | 58.409 | <0.01 | |
| WIL | ASUS1-1H | Monomorphic | ||
| ASUS1-2B | 10 | 44.689 | <0.01 | |
| ASUS1-2F | 10 | 51.854 | <0.01 | |
| ASUS1-3B | 3 | 32.543 | <0.01 | |
| ASUS1-4H | 3 | 41.785 | <0.01 | |
| ASUS1-6C | 28 | 115.403 | <0.01 | |
| ASUS1-8D | 6 | 74.851 | <0.01 | |
| Source | df | SS | MS | Est. Var. | % | θST |
|---|---|---|---|---|---|---|
| Among strains | 2 | 63.978 | 31.989 | 0.893 | 15% | 0.146 (p = 0.001) |
| Within strains | 87 | 453.333 | 5.211 | 5.211 | 85% | |
| Total | 89 | 517.311 | 6.103 | 100% |
| Locus | LAB-TA7 | LAB-WILL | TA7-WILL | Overall |
|---|---|---|---|---|
| ASUS1-1H | 0.015 ns | 0.017 * | 0.115 ns | 0.043 ns |
| ASUS1-2B | 0.033 ns | 0.0 | 0.105 * | 0.049 * |
| ASUS1-1F | 0.239 ** | 0.41 ns | 0.103 ** | 0.158 ** |
| ASUS1-3B | 0.004 ns | 0.028 ns | 0.084 * | 0.044 ns |
| ASUS1-4H | 0.359 ** | 0.389 ** | 0.0 | 0.275 ** |
| ASUS1-6C | 0.030 * | 0.043 | 0.0 | 0.015 ns |
| ASUS1-8D | 0.297 ** | 0.006 | 0.264 * | 0.049 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Montoya, L.; Sánchez-Rosario, M.; López-Gómez, E.; Garcia-Bautista, M.; Canedo-Texón, A.; Haymer, D.; Liedo, P. Mass-Rearing Conditions Do Not Always Reduce Genetic Diversity: The Case of the Mexican Fruit Fly, Anastrepha ludens (Diptera: Tephritidae). Insects 2024, 15, 56. https://doi.org/10.3390/insects15010056
Ruiz-Montoya L, Sánchez-Rosario M, López-Gómez E, Garcia-Bautista M, Canedo-Texón A, Haymer D, Liedo P. Mass-Rearing Conditions Do Not Always Reduce Genetic Diversity: The Case of the Mexican Fruit Fly, Anastrepha ludens (Diptera: Tephritidae). Insects. 2024; 15(1):56. https://doi.org/10.3390/insects15010056
Chicago/Turabian StyleRuiz-Montoya, Lorena, Mayren Sánchez-Rosario, Emiliano López-Gómez, Maricela Garcia-Bautista, Anahí Canedo-Texón, David Haymer, and Pablo Liedo. 2024. "Mass-Rearing Conditions Do Not Always Reduce Genetic Diversity: The Case of the Mexican Fruit Fly, Anastrepha ludens (Diptera: Tephritidae)" Insects 15, no. 1: 56. https://doi.org/10.3390/insects15010056