Male-biased Adult Production of the Striped Fruit Fly, Zeugodacus scutellata, by Feeding dsRNA Specific to Transformer-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Morphological Characters for Sex Determination
2.2. Sequence Analysis
2.3. RNA Extraction and cDNA Preparation
2.4. RT-PCR and RT-qPCR
2.5. Preparation of Double Stranded RNA (dsRNA) with In Vitro Transcription
2.6. Injection or Feeding Treatment of dsRNA to Young Larvae
2.7. Preparation of dsRNA with Recombinant Bacteria
2.8. Quantification of dsRNA Produced from Recombinant Bacteria
2.9. Pretreatment of Recombinant Bacteria Using Heat and Sonication
2.10. Feeding Assay of Recombinant Bacteria Expressing dsRNA
2.11. Statistical Analysis
3. Results
3.1. Morphological Characters of Female and Male Z. scutellata Adults
3.2. Prediction of Transformer-2 (Zs-Tra2) of Z. Scutellata and Expression Profile
3.3. Male-Biased Sex Ratio after RNAi of Zs-Tra2 Expression
3.4. Construction of Recombinant E. coli Expressing dsRNA Specific to Zs-Tra2 and Its Effect on Sex Transformation
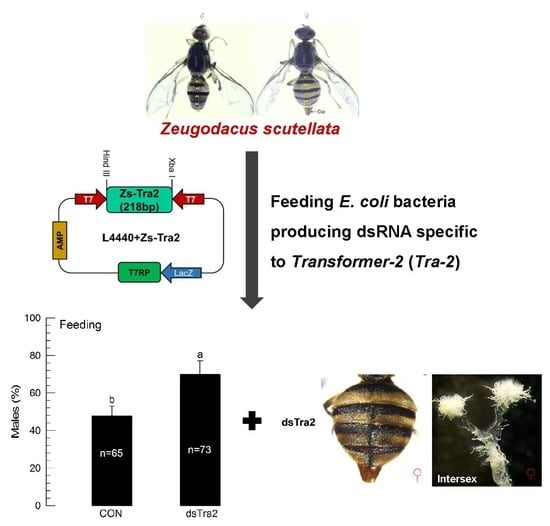
3.5. Effect of Feeding Recombinant E. coli on Sex Ratio of Z. scutellata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vargas, R.I.; Piñero, J.C.; Leblanc, L. An overview of pest species of Bactrocera fruit flies (Diptera: Tephritidae) and the integration of biopesticides with other biological approaches for their management with a focus on the Pacific region. Insects 2015, 6, 297–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korneyev, V. Phylogenetic Relationships among the Families of the Superfamily Tephritoidea. In Fruit Flies (Tephritidae). Phylogeny and Evolution of Behavior; Aluja, M., Norrbom, A., Eds.; CRC: Boca Raton, FL, USA, 1999; pp. 3–22. [Google Scholar]
- Han, H.; Ro, K.-E. Molecular phylogeny of the superfamily Tephritoidea (Insecta: Diptera): New evidence from the mitochondrial 12S, 16S, and COII genes. Mol. Phylogenetics Evol. 2005, 34, 416–430. [Google Scholar] [CrossRef] [PubMed]
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics; CAB International/ACIAR: Boston, MA, USA, 1992. [Google Scholar]
- Schutze, M.K.; Virgilio, M.; Norrbom, A.; Clarke, A.R. Tephritid integrative taxonomy: Where we are now, with a focus on the resolution of three tropical fruit fly species complexes. Annu. Rev. Entomol. 2017, 62, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, D.-S. Integrated pest management against Bactrocera fruit flies. Korean J. Appl. Entomol. 2016, 55, 359–376. [Google Scholar] [CrossRef]
- Han, M.J.; Lee, S.H.; Ahn, S.B.; Choi, J.Y.; Choi, K.M. Distribution, damage and host plants of pumpkin fruit fly, Paradacus depressa (Shiraki). RDA J. Agri. Sci. 1994, 36, 346–350. [Google Scholar]
- Kim, Y.-P.; Jeon, S.-W.; Lee, S.-G.; Kim, K.-H.; Choi, N.-J.; Hwang, C.-Y. Seasonal occurrence and damage of Bactrocera scutellata (Diptera: Tephritidae) in Jeonbuk province. Korean J. Appl. Entomol. 2010, 49, 299–304. [Google Scholar] [CrossRef]
- Shiraki, T. Fruit flies of the Ryukyu islands. Bull. United States Natl. Mus. 1968, 263, 1–104. [Google Scholar] [CrossRef]
- Miyatake, T.; Kuba, H.; Yukawa, J. Seasonal occurrence of Bactrocera scutellata (Diptera: Tephritidae), a cecidophage of stem galls produced by Lasioptera sp. (Diptera: Cecidomyiidae) on wild gourds (Cucurbitaceae). Ann. Entomol. Soc. Am. 2000, 93, 1274–1279. [Google Scholar] [CrossRef]
- Ohno, S.; Haraguchi, D.; Kohama, T. New host and distribution records of the fruit fly, Bactrocera scutellata (Hendel) (Diptera: Tephritidae), in Southwestern Japan, and a case of infestation of the species on cucumber fruits at Okinawa Island. Jpn. J. Entomol. 2006, 9, 7–9. [Google Scholar]
- Enkerlin, W.R. Impact of fruit fly control programs using the sterile insect technique. In Sterile Insect Technique Principles and Practice in Area-Wide Integrated Pest Management; Dyck, V.A., Hendrichs, J., Robinson, A.S., Eds.; Springer: Berlin, Germany, 2005; pp. 651–676. [Google Scholar]
- Knipling, E.F. Possibilities of Insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 1955, 48, 459–462. [Google Scholar] [CrossRef]
- Rendón, P.; McInnis, D.; Lance, D.; Stewart, J. Medfly (Diptera: Tephritidae) genetic sexing: Large-scale field comparison of males-only and bisexual sterile fly releases in Guatemala. J. Econ. Entomol. 2004, 97, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- McInnis, D.; Tam, S.; Lim, R.; Komatsu, J.; Kurashima, R.; Albrecht, C. Development of a pupal color-based genetic sexing strain of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 2004, 97, 1026–1033. [Google Scholar] [CrossRef] [Green Version]
- Ogaugwu, C.E.; Schetelig, M.F.; Wimmer, E.A. Transgenic sexing system for Ceratitis capitata (Diptera: Tephritidae) based on female-specific embryonic lethality. Insect Biochem. Mol. Boil. 2013, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cline, T.W. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993, 9, 385–390. [Google Scholar] [CrossRef]
- Sharma, A.; Heinze, S.D.; Wu, Y.; Kohlbrenner, T.; Morilla, I.; Brunner, C.; Wimmer, E.A.; Van De Zande, L.; Robinson, M.D.; Beukeboom, L.W.; et al. Male sex in houseflies is determined byMdmd, a paralog of the generic splice factor geneCWC22. Science 2017, 356, 642–645. [Google Scholar] [CrossRef]
- Salvemini, M.; Robertson, M.; Aronson, B.; Atkinson, P.; Polito, L.C.; Saccone, G. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Boil. 2009, 53, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Wu, Q.; Li, J.-W.; Zhang, G.; Wan, F.-H. RNAi-mediated knock-down of transformer and transformer 2 to generate male-only progeny in the Oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS ONE 2015, 10, e0128892. [Google Scholar] [CrossRef] [Green Version]
- Thongsaiklaing, T.; Nipitwattanaphon, M.; Ngernsiri, L. The transformer2 gene of the pumpkin fruit fly, Bactrocera tau (Walker), functions in sex determination, male fertility and testis development. Insect Mol. Boil. 2018, 27, 766–779. [Google Scholar] [CrossRef]
- Bustin, S.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Park, B.; Kim, Y. Transient transcription of a putative RNase containing BEN domain encoded in Cotesia plutellae bracovirus induces an immunosuppression of the diamondback moth, Plutella xylostella. J. Invertebr. Pathol. 2010, 105, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Park, Y.; Kim, Y. A transformed bacterium expressing double-stranded RNA specific to integrin β1 enhances Bt toxin efficacy against a polyphagous insect pest, Spodoptera exigua. PLoS ONE 2015, 10, e0132631. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS/STAT User’s Guide; SAS Institute: Cary, NC, USA, 1989. [Google Scholar]
- Jeon, S.-W.; Kang, T.-J.; Cho, M.-R.; Kim, K.-H.; Lee, S.G.; Kim, J.S.; Park, H.W. Adult longevity and life table analysis of striped fruit fly, Bactrocera scutellata (Hendel) (Diptera:Tephritidae). Korean J. Appl. Entomol. 2012, 51, 485–488. [Google Scholar] [CrossRef]
- Kim, K.; Kim, M.; Kwon, G.; Kim, Y. Technologies required for development of trap-based MAT control against the striped fruit fly, Bactrocera scutellata. Korean J. Appl. Entomol. 2017, 56, 51–60. [Google Scholar] [CrossRef]
- Kim, Y.; Al Baki, M.A.; Kwon, G. Technique to generate sterile males of striped fruit flies, Zeugodacus scutellata, using electron beam irradiation and their application to genetic control. Korean J. Appl. Entomol. 2020, 59, 1–10. [Google Scholar]
- Amrein, H.; Maniatis, T.; Nöthiger, R. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. EMBO J. 1990, 9, 3619–3629. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.S.; Wolfner, M.F. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genome Res. 1988, 2, 477–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshijima, K.; Inoue, K.; Higuchi, I.; Sakamoto, H.; Shimura, Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 1991, 252, 833–836. [Google Scholar] [CrossRef]
- Mattox, W.; Palmer, M.J.; Baker, B.S. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 1990, 4, 789–805. [Google Scholar] [CrossRef] [Green Version]
- Mattox, W.; Baker, B.S. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991, 5, 786–796. [Google Scholar] [CrossRef] [Green Version]
- Mattox, W.; McGuffin, M.E.; Baker, B.S. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics 1996, 143, 303–314. [Google Scholar] [PubMed]
- McGuffin, M.E.; Chandler, D.; Somaiya, D.; Dauwalder, B.; Mattox, W. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics 1998, 149, 1477–1486. [Google Scholar] [PubMed]
- Sarno, F.; Ruiz, M.F.; Eirin-Lopez, J.M.; Perondini, A.; Selivon, D.; Sánchez, L. The gene transformer-2 of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. BMC Evol. Boil. 2010, 10, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schetelig, M.F.; Milano, A.; Saccone, G.; Handler, A.M. Male only progeny in Anastrepha suspensa by RNAi-induced sex reversion of chromosomal females. Insect Biochem. Mol. Boil. 2012, 42, 51–57. [Google Scholar] [CrossRef]
- Lagos, D.; Koukidou, M.; Savakis, C.; Komitopoulou, K. The transformer gene in Bactrocera oleae: The genetic switch that determines its sex fate. Insect Mol. Boil. 2007, 16, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.L.; Riegler, M.; Frommer, M.; Shearman, D. Expression patterns of sex-determination genes in single male and female embryos of two Bactrocera fruit fly species during early development. Insect Mol. Boil. 2014, 23, 754–767. [Google Scholar] [CrossRef]
- Pane, A.; Salvemini, M.; Delli Bovi, P.; Polito, C.; Saccone, G. Faculty of 1000 evaluation for The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 2002, 129, 3715–3725. [Google Scholar]
- Hediger, M.; Siegenthaler, C.; Moser, M.; Bopp, D. The transformer2 gene in Musca domestica is required for selecting and maintaining the female pathway of development. Dev. Genes Evol. 2005, 215, 165–176. [Google Scholar]
- Liu, J.; Smagghe, G.; Swevers, L. Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J. Insect Physiol. 2013, 59, 646–654. [Google Scholar] [CrossRef]
- Allen, M.; Walker, W.B.; Iii, W.B.W. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Baki, M.A.; Vatanparast, M.; Kim, Y. Male-biased Adult Production of the Striped Fruit Fly, Zeugodacus scutellata, by Feeding dsRNA Specific to Transformer-2. Insects 2020, 11, 211. https://doi.org/10.3390/insects11040211
Al Baki MA, Vatanparast M, Kim Y. Male-biased Adult Production of the Striped Fruit Fly, Zeugodacus scutellata, by Feeding dsRNA Specific to Transformer-2. Insects. 2020; 11(4):211. https://doi.org/10.3390/insects11040211
Chicago/Turabian StyleAl Baki, Md. Abdullah, Mohammad Vatanparast, and Yonggyun Kim. 2020. "Male-biased Adult Production of the Striped Fruit Fly, Zeugodacus scutellata, by Feeding dsRNA Specific to Transformer-2" Insects 11, no. 4: 211. https://doi.org/10.3390/insects11040211