Inhibitory Efficacy of Main Components of Scutellaria baicalensis on the Interaction between Spike Protein of SARS-CoV-2 and Human Angiotensin-Converting Enzyme II
Abstract
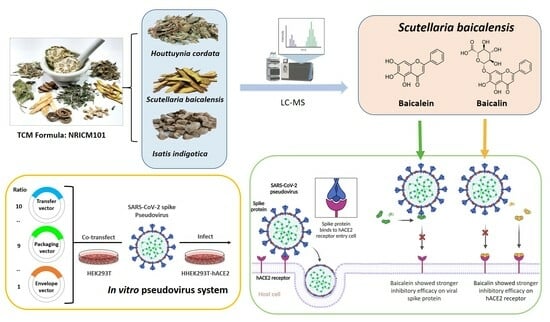
:1. Introduction
2. Results
2.1. Establishment of a Pseudovirus System for Evaluating the Binding Efficacy of Viral Spike Protein and hACE2 Receptor
2.2. Identification of SARS-CoV-2 Spike Pseudovirus System for Drug Candidates
2.3. Assessment of Inhibitory Efficacy of Herbs on Pseudovirus Infection
2.4. Identification of Active Ingredients in S. baicalensis
2.5. Baicalein and Baicalin Inhibition of Omicron Spike Pseudovirus Infection
2.6. Synergistic Effect of Baicalein and Baicalin on Pseudovirus Infection Inhibition
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Cell Viability Assay
4.3. Pseudovirus System
4.4. Pseudovirus Infection Inhibition Assay
4.5. Protein Extraction
4.6. Western Blot
4.7. Extraction of Each TCM
4.8. LC-MS Analyses
4.9. Synergy Scoring and Detection
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Beyond Omicron: What’s next for COVID’s viral evolution. Nature 2021, 600, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Pustake, M.; Tambolkar, I.; Giri, P.; Gandhi, C. SARS, MERS and CoVID-19: An overview and comparison of clinical, laboratory and radiological features. J. Family Med. Prim. Care 2022, 11, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Saldivar-Espinoza, B.; Garcia-Segura, P.; Novau-Ferré, N.; Macip, G.; Martínez, R.; Puigbò, P.; Cereto-Massagué, A.; Pujadas, G.; Garcia-Vallve, S. The Mutational Landscape of SARS-CoV-2. Int. J. Mol. Sci. 2023, 24, 9072. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. A detailed overview of SARS-CoV-2 omicron: Its sub-variants, mutations and pathophysiology, clinical characteristics, immunological landscape, immune escape, and therapies. Viruses 2023, 15, 167. [Google Scholar] [CrossRef]
- Lamkiewicz, K.; Esquivel Gomez, L.R.; Kühnert, D.; Marz, M. Genome structure, life cycle, and taxonomy of coronaviruses and the evolution of SARS-CoV-2. In Viral Fitness and Evolution: Population Dynamics and Adaptive Mechanisms; Springer: Berlin/Heidelberg, Germany, 2023; pp. 305–339. [Google Scholar]
- Hirano, T.; Murakami, M. COVID-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef]
- Yang, L.; Li, J.; Guo, S.; Hou, C.; Liao, C.; Shi, L.; Ma, X.; Jiang, S.; Zheng, B.; Fang, Y. SARS-CoV-2 variants, RBD mutations, binding affinity, and antibody escape. Int. J. Mol. Sci. 2021, 22, 12114. [Google Scholar] [CrossRef]
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): A structural perspective. Mol. Biol. Rep. 2023, 50, 2713–2721. [Google Scholar] [CrossRef]
- Reuter, N.; Chen, X.; Kropff, B.; Peter, A.S.; Britt, W.J.; Mach, M.; Überla, K.; Thomas, M. SARS-CoV-2 Spike Protein Is Capable of Inducing Cell–Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies. Viruses 2023, 15, 1500. [Google Scholar] [CrossRef]
- Marshall, A.C. Traditional Chinese Medicine and Clinical Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 455–482. [Google Scholar]
- Lyu, M.; Fan, G.; Xiao, G.; Wang, T.; Xu, D.; Gao, J.; Ge, S.; Li, Q.; Ma, Y.; Zhang, H. Traditional Chinese medicine in COVID-19. Acta Pharm. Sin. B 2021, 11, 3337–3363. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, Y.; Zhou, L.; Zhou, X.; Xie, B.; Zhang, W.; Sun, J. Prevention and treatment of COVID-19 using Traditional Chinese Medicine: A review. Phytomedicine 2021, 85, 153308. [Google Scholar] [CrossRef]
- Martín Giménez, V.M.; Modrego, J.; Gómez-Garre, D.; Manucha, W.; de Las Heras, N. Gut microbiota dysbiosis in COVID-19: Modulation and approaches for prevention and therapy. Int. J. Mol. Sci. 2023, 24, 12249. [Google Scholar] [CrossRef]
- Tsai, K.C.; Huang, Y.C.; Liaw, C.C.; Tsai, C.I.; Chiou, C.T.; Lin, C.J.; Wei, W.C.; Lin, S.J.; Tseng, Y.H.; Yeh, K.M.; et al. A traditional Chinese medicine formula NRICM101 to target COVID-19 through multiple pathways: A bedside-to-bench study. Biomed. Pharmacother. 2021, 133, 111037. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Peng, W.Y.; Lee, W.H.; Lin, T.Y.; Yang, M.H.; Dalley, J.W.; Tsai, T.H. Biotransformation and brain distribution of the anti-COVID-19 drug molnupiravir and herb-drug pharmacokinetic interactions between the herbal extract Scutellaria formula-NRICM101. J. Pharm. Biomed. Anal. 2023, 115499. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Chen, Y.J.; Lin, M.W.; Chang, H.J.; Yang, X.R.; Lin, C.S. ACE2 and a Traditional Chinese Medicine Formula NRICM101 Could Alleviate the Inflammation and Pathogenic Process of Acute Lung Injury. Medicina 2023, 59, 1554. [Google Scholar] [CrossRef]
- Wang, M.; Tao, L.; Xu, H. Chinese herbal medicines as a source of molecules with anti-enterovirus 71 activity. Chin. Med. 2016, 11, 2. [Google Scholar] [CrossRef]
- Zhao, T.; Tang, H.; Xie, L.; Zheng, Y.; Ma, Z.; Sun, Q.; Li, X. Scutellaria baicalensis Georgi. (Lamiaceae): A review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J. Pharm. Pharmacol. 2019, 71, 1353–1369. [Google Scholar] [CrossRef]
- Chen, Q.; Lan, H.Y.; Peng, W.; Rahman, K.; Liu, Q.C.; Luan, X.; Zhang, H. Isatis indigotica: A review of phytochemistry, pharmacological activities and clinical applications. J. Pharm. Pharmacol. 2021, 73, 1137–1150. [Google Scholar] [CrossRef]
- Kaul, R.; Paul, P.; Kumar, S.; Büsselberg, D.; Dwivedi, V.D.; Chaari, A. Promising antiviral activities of natural flavonoids against SARS-CoV-2 targets: Systematic review. Int. J. Mol. Sci. 2021, 22, 11069. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Depicting the inhibitory potential of polyphenols from Isatis indigotica root against the main protease of SARS CoV-2 using computational approaches. J. Biomol. Struct. Dyn. 2022, 40, 4110–4121. [Google Scholar] [CrossRef] [PubMed]
- Thoene, J.; Gavin, R.F.; Towne, A.; Wattay, L.; Ferrari, M.G.; Navarrete, J.; Pal, R. In vitro activity of cysteamine against SARS-CoV-2 variants. Mol. Genet. Metab. 2022, 137, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Alonzi, T.; Aiello, A.; Repele, F.; Falasca, L.; Francalancia, M.; Garbuglia, A.R.; Delogu, G.; Nicastri, E.; Piacentini, M.; Goletti, D.; et al. Cysteamine exerts in vitro antiviral activity against the SARS-CoV-2 Delta and Omicron variants. Cell Death Discov. 2020, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yang, M.L.; Duan, Z.L.; Liu, F.L.; Jin, L.; Long, C.B.; Zhang, M.; Tang, X.P.; Xu, L.; Li, Y.C.; et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021, 31, 17–24. [Google Scholar] [CrossRef]
- Hoffmann, M.; Jin, Y.; Pöhlmann, S. Dalbavancin: Novel candidate for COVID-19 treatment. Cell Res. 2021, 31, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Yu, M.; Shi, M.; Bo, P.; Gu, X.; Zhang, Z. Baicalin and its aglycone: A novel approach for treatment of metabolic disorders. Pharmacol. Rep. 2020, 72, 13–23. [Google Scholar] [CrossRef]
- Liang, S.; Deng, X.; Lei, L.; Zheng, Y.; Ai, J.; Chen, L.; Xiong, H.; Mei, Z.; Cheng, Y.-C.; Ren, Y. The comparative study of the therapeutic effects and mechanism of baicalin, baicalein, and their combination on ulcerative colitis rat. Front. Pharmacol. 2019, 10, 1466. [Google Scholar] [CrossRef]
- Dinda, B.; Dinda, S.; DasSharma, S.; Banik, R.; Chakraborty, A.; Dinda, M. Therapeutic potentials of baicalin and its aglycone, baicalein against inflammatory disorders. Eur. J. Med. Chem. 2017, 131, 68–80. [Google Scholar] [CrossRef]
- Song, J.W.; Long, J.Y.; Xie, L.; Zhang, L.L.; Xie, Q.X.; Chen, H.J.; Deng, M.; Li, X.F. Applications, phytochemistry, pharmacological effects, pharmacokinetics, toxicity of Scutellaria baicalensis Georgi. and its probably potential therapeutic effects on COVID-19: A review. Chin. Med. 2020, 15, 102. [Google Scholar] [CrossRef]
- Boozari, M.; Hosseinzadeh, H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytother. Res. 2021, 35, 864–876. [Google Scholar] [CrossRef]
- Jia, K.; Li, Y.; Liu, T.; Gu, X.; Li, X. New insights for infection mechanism and potential targets of COVID-19: Three Chinese patent medicines and three Chinese medicine formulas as promising therapeutic approaches. Chin. Herb. Med. 2023, 15, 157–168. [Google Scholar] [CrossRef]
- Fuzimoto, A.D.; Isidoro, C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds-Additional weapons in the fight against the COVID-19 pandemic? J. Tradit. Complement. Med. 2020, 10, 405–419. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef]
- Chun, J.M.; Kim, H.S.; Lee, A.Y.; Kim, S.H.; Kim, H.K. Anti-Inflammatory and Antiosteoarthritis Effects of Saposhnikovia divaricata ethanol Extract: In Vitro and In Vivo Studies. Evid.-Based Complement. Altern. Med. 2016, 2016, 1984238. [Google Scholar]
- Laldinsangi, C. The therapeutic potential of Houttuynia cordata: A current review. Heliyon 2022, 8, e10386. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Han, Y.; Wang, W.; Liang, G.; Qi, J. Mulberry leaves attenuate D-galactose-induced aging in vivo and in vitro. J. Ethnopharmacol. 2023, 311, 116286. [Google Scholar]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Dinda, B.; Dinda, M.; Dinda, S.; De, U.C. An overview of anti-SARS-CoV-2 and anti-inflammatory potential of baicalein and its metabolite baicalin: Insights into molecular mechanisms. Eur. J. Med. Chem. 2023, 258, 115629. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Snyder, S.A.; Smith, J.N.; Chen, Y.C. Anticancer properties of baicalein: A review. Med. Chem. Res. 2016, 25, 1515–1523. [Google Scholar] [CrossRef]
- Yang, J.Y.; Ma, Y.X.; Liu, Y.; Peng, X.J.; Chen, X.Z. A Comprehensive Review of Natural Flavonoids with Anti-SARS-CoV-2 Activity. Molecules 2023, 28, 2735. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Tsai, F.J.; Hsu, Y.M.; Ho, T.J.; Wang, G.K.; Chiu, Y.J.; Ha, H.A.; Yang, J.S. Study of Baicalin toward COVID-19 Treatment: In silico Target Analysis and in vitro Inhibitory Effects on SARS-CoV-2 Proteases. Biomed. Hub. 2021, 6, 122–137. [Google Scholar] [CrossRef]
- Zandi, K.; Musall, K.; Oo, A.; Cao, D.; Liang, B.; Hassandarvish, P.; Lan, S.; Slack, R.L.; Kirby, K.A.; Bassit, L.; et al. Baicalein and Baicalin Inhibit SARS-CoV-2 RNA-Dependent-RNA Polymerase. Microorganisms 2021, 9, 893. [Google Scholar] [CrossRef]
- Feng, J.; Li, D.; Zhang, J.; Yin, X.; Li, J. Crystal structure of SARS-CoV 3C-like protease with baicalein. Biochem. Biophys. Res. Commun. 2022, 611, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Su, H.; Ke, C.; Tang, C.; Witt, M.; Quinn, R.J.; Xu, Y.; Liu, J.; Ye, Y. Efficient discovery of potential inhibitors for SARS-CoV-2 3C-like protease from herbal extracts using a native MS-based affinity-selection method. J. Pharm. Biomed. Anal. 2022, 209, 114538. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, J. Current findings regarding natural components with potential anti-2019-nCoV activity. Front. Cell Dev. Biol. 2020, 8, 589. [Google Scholar] [CrossRef]
- Song, J.B.; Zhao, L.Q.; Wen, H.P.; Li, Y.P. Herbal combinations against COVID-19: A network pharmacology, molecular docking and dynamics study. J. Integr. Med. 2023, 21, 593–604. [Google Scholar] [CrossRef]
- Shakhsi-Niaei, M.; Soureshjani, E.H.; Babaheydari, A.K. In silico comparison of separate or combinatorial effects of potential inhibitors of the SARS-CoV-2 binding site of ACE2. Iran. J. Public Health 2021, 50, 1028. [Google Scholar] [CrossRef]
- KalantarMotamedi, Y.; Eastman, R.T.; Guha, R.; Bender, A. A systematic and prospectively validated approach for identifying synergistic drug combinations against malaria. Malar. J. 2018, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Giri, A.K.; Gautam, P.; Kononov, A.; Potdar, S.; Saarela, J.; Wennerberg, K.; Aittokallio, T. Prediction of drug combination effects with a minimal set of experiments. Nat. Mach. Intell. 2019, 1, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; He, L.; Aittokallio, T.; Tang, J. SynergyFinder: A web application for analyzing drug combination dose–response matrix data. Bioinformatics 2017, 33, 2413–2415. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Wennerberg, K.; Aittokallio, T.; Tang, J. Searching for drug synergy in complex dose–response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2015, 13, 504–513. [Google Scholar] [CrossRef]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020, 48, W488–W493. [Google Scholar] [CrossRef]








| Groups | Treatment |
|---|---|
| SPV | Spike blocker (Cysteamine) pre-treated with SARS-CoV-2 spike pseudovirus |
| SPC | Spike blocker pre-treated with HEK293T-hACE2 cells |
| APV | hACE2 blocker (Dalbavancin) pre-treated with SARS-CoV-2 spike pseudovirus |
| APC | hACE2 blocker pre-treated with HEK293T-hACE2 cells |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-H.; Chang, H.-J.; Lin, M.-W.; Yang, X.-R.; Lee, C.-H.; Lin, C.-S. Inhibitory Efficacy of Main Components of Scutellaria baicalensis on the Interaction between Spike Protein of SARS-CoV-2 and Human Angiotensin-Converting Enzyme II. Int. J. Mol. Sci. 2024, 25, 2935. https://doi.org/10.3390/ijms25052935
Lin C-H, Chang H-J, Lin M-W, Yang X-R, Lee C-H, Lin C-S. Inhibitory Efficacy of Main Components of Scutellaria baicalensis on the Interaction between Spike Protein of SARS-CoV-2 and Human Angiotensin-Converting Enzyme II. International Journal of Molecular Sciences. 2024; 25(5):2935. https://doi.org/10.3390/ijms25052935
Chicago/Turabian StyleLin, Cheng-Han, Ho-Ju Chang, Meng-Wei Lin, Xin-Rui Yang, Che-Hsiung Lee, and Chih-Sheng Lin. 2024. "Inhibitory Efficacy of Main Components of Scutellaria baicalensis on the Interaction between Spike Protein of SARS-CoV-2 and Human Angiotensin-Converting Enzyme II" International Journal of Molecular Sciences 25, no. 5: 2935. https://doi.org/10.3390/ijms25052935