Tumor-Promoting Activity and Proteomic Profiling of Cisplatin/Oxaliplatin-Derived DAMPs in Cholangiocarcinoma Cells
Abstract
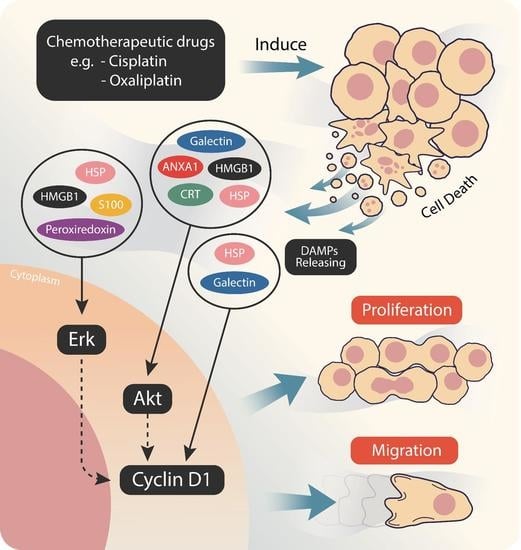
:1. Introduction
2. Results
2.1. Cisplatin and Oxaliplatin Caused CCA Cell Toxicity
2.2. Cisplatin and Oxaliplatin Induced DAMP Expression in CCA Cells
2.3. Chemo-Drug-Derived DAMPs Promoted CCA Cells’ Proliferation and Migration
2.4. Chemo-Drug-Derived DAMPs Activated the Pro-Survival Signal of CCA Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MTT Assay
4.3. Annexin V/PI Staining
4.4. LDH Activity Measurement
4.5. Apoptosis Protein Measurement
4.6. Preparation of Chemo-Drug-Derived DAMPs
4.7. In-Solution Protein Digestion
4.8. Label-Free Nano-LC-MS/MS Analysis
4.9. MS Data Processing and Statistical Analysis
4.10. Enzyme-Linked Immunosorbent Assay (ELISA)
4.11. ATP Measurement
4.12. Calreticulin Measurement
4.13. Cell Migration Assay
4.14. Immunoblotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, K.; Zhao, R.; Ji, T.; Wang, X.; Yang, X.; Zhang, Y.; Cheng, K.; Liu, S.; Hao, J.; et al. Inducing enhanced immunogenic cell death with nanocarrier-based drug delivery systems for pancreatic cancer therapy. Biomaterials 2016, 102, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Ye, W.; Xiao, R.; Silvin, C.; Padget, M.; Hodge, J.W.; Van Waes, C.; Schmitt, N.C. Cisplatin and oxaliplatin induce similar immunogenic changes in preclinical models of head and neck cancer. Oral Oncol. 2019, 95, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Aoto, K.; Mimura, K.; Okayama, H.; Saito, M.; Chida, S.; Noda, M.; Nakajima, T.; Saito, K.; Abe, N.; Ohki, S.; et al. Immunogenic tumor cell death induced by chemotherapy in patients with breast cancer and esophageal squamous cell carcinoma. Oncol. Rep. 2018, 39, 151–159. [Google Scholar] [CrossRef]
- Casares, N.; Pequignot, M.O.; Tesniere, A.; Ghiringhelli, F.; Roux, S.; Chaput, N.; Schmitt, E.; Hamai, A.; Hervas-Stubbs, S.; Obeid, M.; et al. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J. Exp. Med. 2005, 202, 1691–1701. [Google Scholar] [CrossRef]
- Economopoulou, P.; Koutsodontis, G.; Strati, A.; Kirodimos, E.; Giotakis, E.; Maragoudakis, P.; Prikas, C.; Papadimitriou, N.; Perisanidis, C.; Gagari, E.; et al. Surrogates of immunologic cell death (ICD) and chemoradiotherapy outcomes in head and neck squamous cell carcinoma (HNSCC). Oral Oncol. 2019, 94, 93–100. [Google Scholar] [CrossRef]
- Krysko, O.; Aaes, T.L.; Bachert, C.; Vandenabeele, P.; Krysko, D.V. Many faces of DAMPs in cancer therapy. Cell Death Dis. 2013, 4, e631. [Google Scholar] [CrossRef]
- He, S.; Cheng, J.; Sun, L.; Wang, Y.; Wang, C.; Liu, X.; Zhang, Z.; Zhao, M.; Luo, Y.; Tian, L.; et al. HMGB1 released by irradiated tumor cells promotes living tumor cell proliferation via paracrine effect. Cell Death Dis. 2018, 9, 648. [Google Scholar] [CrossRef]
- Tripathi, A.; Shrinet, K.; Kumar, A. HMGB1 protein as a novel target for cancer. Toxicol. Rep. 2019, 6, 253–261. [Google Scholar] [CrossRef]
- Anunobi, R.; Boone, B.A.; Cheh, N.; Tang, D.; Kang, R.; Loux, T.; Lotze, M.T.; Zeh, H.J. Extracellular DNA promotes colorectal tumor cell survival after cytotoxic chemotherapy. J. Surg. Res. 2018, 226, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, C.; Huebener, P.; Schwabe, R.F. Damage-associated molecular patterns in cancer: A double-edged sword. Oncogene 2016, 35, 5931–5941. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jing, J.; Ye, Y.; Chen, Z.; Jing, Y.; Li, S.; Hong, W.; Ruan, H.; Liu, Y.; Hu, Q.; et al. Characterization of the dual functional effects of heat shock proteins (HSPs) in cancer hallmarks to aid development of HSP inhibitors. Genome Med. 2020, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Sang, L.; Wang, X. S100 Calcium Binding Protein A11 (S100A11) Promotes The Proliferation, Migration and Invasion of Cervical Cancer Cells, and Activates Wnt/beta-Catenin Signaling. Onco Targets Ther. 2019, 12, 8675–8685. [Google Scholar] [CrossRef] [PubMed]
- Juan, V.H.W.; Daniel, H.P.; David, C.; Alan, A.; Anthony, M.; Srinivasan, M.; Tim, I.; Sharon, H.; Stephen, P.P.; Michael, R.; et al. Cisplatin plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar]
- Jang, J.S.; Lim, H.Y.; Hwang, I.G.; Song, H.S.; Yoo, N.; Yoon, S.; Kim, Y.H.; Park, E.; Byun, J.H.; Lee, M.A.; et al. Gemcitabine and oxaliplatin in patients with unresectable biliary cancer including gall bladder cancer: A Korean Cancer Study Group phase II trial. Cancer Chemother. Pharmacol. 2010, 65, 641–647. [Google Scholar] [CrossRef]
- Solari, J.I.G.; Filippi-Chiela, E.; Pilar, E.S.; Nunes, V.; Gonzalez, E.A.; Figueiro, F.; Andrade, C.F.; Klamt, F. Damage-associated molecular patterns (DAMPs) related to immunogenic cell death are differentially triggered by clinically relevant chemotherapeutics in lung adenocarcinoma cells. BMC Cancer 2020, 20, 474. [Google Scholar] [CrossRef]
- Stojanovska, V.; Prakash, M.; McQuade, R.; Fraser, S.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Oxaliplatin Treatment Alters Systemic Immune Responses. BioMed Res. Int. 2019, 2019, 4650695. [Google Scholar] [CrossRef]
- Murao, A.; Aziz, M.; Wang, H.; Brenner, M.; Wang, P. Release mechanisms of major DAMPs. Apoptosis 2021, 26, 152–162. [Google Scholar] [CrossRef]
- Radic, M.; Marion, T.; Monestier, M. Nucleosomes are exposed at the cell surface in apoptosis. J. Immunol. 2004, 172, 6692–6700. [Google Scholar] [CrossRef]
- Jiang, W.; Bell, C.W.; Pisetsky, D.S. The relationship between apoptosis and high-mobility group protein 1 release from murine macrophages stimulated with lipopolysaccharide or polyinosinic-polycytidylic acid. J. Immunol. 2007, 178, 6495–6503. [Google Scholar] [CrossRef]
- Chen, R.; Kang, R.; Fan, X.G.; Tang, D. Release and activity of histone in diseases. Cell Death Dis. 2014, 5, e1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Binder, J.R.; Suto, R.; Anderson, M.K.; Srivastana, K.P. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int. Immunol. 2000, 12, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Strzyz, P. Cell death: Pulling the apoptotic trigger for necrosis. Nat. Rev. Mol. Cell Biol. 2017, 18, 72. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Z.; Yang, G.W.; Li, H.; Chen, Q.; Song, R.; Zhao, L. HMGB1 overexpression as a prognostic factor for survival in cancer: A meta-analysis and systematic review. Oncotarget 2016, 7, 50417–50427. [Google Scholar] [CrossRef]
- Albakova, Z.; Siam, M.K.S.; Sacitharan, P.K.; Ziganshin, R.H.; Ryazantsev, D.Y.; Sapozhnikov, A.M. Extracellular heat shock proteins and cancer: New perspectives. Transl. Oncol. 2021, 14, 100995. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Yan, Y.; Guo, L.; Tian, F.; Wu, D. Prognostic role of HSPs in human gastrointestinal cancer: A systematic review and meta-analysis. Onco Targets Ther. 2018, 11, 351–359. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, 4. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Rouhiainen, A.; Kuja-Panula, J.; Tumova, S.; Rauvala, H. RAGE-mediated cell signaling. Methods Mol. Biol. 2013, 963, 239–263. [Google Scholar] [CrossRef]
- Yan, S.F.; Ramasamy, R.; Schmidt, A.M. The RAGE axis: A fundamental mechanism signaling danger to the vulnerable vasculature. Circ. Res. 2010, 106, 842–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Landi, F.; Bernabei, R.; Romano, R.; Bucci, C.; Marzetti, E. Extracellular vesicles and damage-associated molecular patterns: A Pandora’s box in health and disease. Front. Immunol. 2020, 11, 601740. [Google Scholar] [CrossRef] [PubMed]






| Accession No. | Protein Name | MW (kDa) | Quantitative Intensity | Fold-Change | p-Value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Control | CIS | OXA | CIS | OXA | CIS | OXA | |||
| P61604 | 10 kDa heat shock protein, mitochondrial | 11 | 15,427 | 198,630 | 482,330 | 12.88 | 31.27 | 0.0009 | 0.0001 |
| P10809 | 60 kDa heat shock protein, mitochondrial | 61 | 298,183 | 10,427,833 | 6,603,367 | 34.97 | 22.15 | 0.0009 | 0.0035 |
| P04083 | Annexin A1 | 39 | 709,853 | 620,593 | 2,785,267 | 0.87 | 3.92 | 0.4866 | 0.0038 |
| P07355 | Annexin A2 | 39 | 159,363 | 1,715,067 | 2,038,033 | 10.76 | 12.79 | 0.0020 | 0.0067 |
| P12429 | Annexin A3 | 36 | 1,748,067 | 2,358,433 | 1,273,867 | 1.35 | 0.73 | 0.1356 | 0.0970 |
| V9M9E1 | ATP synthase protein 8 | 8 | 1435 | 17,277 | 2138 | 12.04 | 1.49 | 0.0015 | 0.2006 |
| Q9H1M4 | Beta-defensin 127 | 11 | 14,510 | 2,656,900 | 1,967,767 | 183.11 | 135.61 | 0.0014 | 0.0009 |
| A6NLG9 | Biglycan | 35 | 11,992 | 2,858,133 | 2,728,933 | 238.34 | 227.57 | 0.0001 | 0.0004 |
| P27797 | Calreticulin | 48 | 71,323 | 2,508,657 | 1,840,800 | 35.17 | 25.81 | 0.0850 | 0.0014 |
| Q96L12 | Calreticulin-3 | 45 | 123,947 | 1,659,800 | 1,481,167 | 13.39 | 11.95 | 0.0000 | 0.0007 |
| Q05315 | Galectin-10 | 16 | 2819 | 476,200 | 599,833 | 168.91 | 212.76 | 0.0004 | 0.0009 |
| Q96DT0 | Galectin-12 | 38 | 527,307 | 874,453 | 530,443 | 1.66 | 1.01 | 0.0100 | 0.9642 |
| P17931 | Galectin-3 | 26 | 40 | 286,617 | 451,240 | 7165.42 | 11,281 | 0.0001 | 0.0005 |
| P56470 | Galectin-4 | 36 | 5787 | 481,857 | 600,410 | 83.26 | 103.75 | 0.0002 | 0.0001 |
| O00214 | Galectin-8 | 36 | 82,826 | 1,330,867 | 681,423 | 16.07 | 8.23 | 0.0001 | 0.0012 |
| Q6DKI2 | Galectin-9C | 40 | 40 | 641,557 | 474,763 | 16,038.92 | 11,869.08 | 0.0001 | 0.0031 |
| A0A024R4D5 | Glypican-1 | 54 | 10,264 | 1,750,100 | 2,619,967 | 170.51 | 255.25 | 0.0001 | 0.0029 |
| A0A0J9YXG7 | Glypican-2 | 14 | 88,987 | 1,760,067 | 1,756,433 | 19.78 | 19.74 | 0.0001 | 0.0019 |
| Q8IYG2 | Glypican-3 | 66 | 1764 | 84,857 | 118,769 | 48.09 | 67.31 | 0.0018 | 0.0007 |
| A0A087WX91 | Glypican-5 | 36 | 28,399 | 1,273,533 | 565,997 | 44.84 | 19.93 | 0.0004 | 0.0023 |
| Q9Y625 | Glypican-6; Secreted glypican-6 | 63 | 107,510 | 3,483,800 | 2,420,267 | 32.40 | 22.51 | 0.0003 | 0.0003 |
| P48723 | Heat shock 70 kDa protein 13 | 52 | 36,146 | 1,438,600 | 775,427 | 39.80 | 21.45 | 0.0005 | 0.0856 |
| Q0VDF9 | Heat shock 70 kDa protein 14 | 55 | 3834 | 1,665,500 | 2,451,500 | 434.40 | 639.41 | 0.0032 | 0.0006 |
| P0DMV8 | Heat shock 70 kDa protein 1A | 70 | 70,951 | 1,216,900 | 1,271,600 | 17.15 | 17.92 | 0.0001 | 0.0006 |
| P0DMV9 | Heat shock 70 kDa protein 1B | 70 | 361,193 | 3,071,400 | 2,968,700 | 8.50 | 8.22 | 0.0007 | 0.0032 |
| P34932 | Heat shock 70 kDa protein 4 | 94 | 941,070 | 4,376,200 | 3,807,100 | 4.65 | 4.05 | 0.0016 | 0.0032 |
| Q00613 | Heat shock factor protein 1 | 57 | 45,915 | 1,332,633 | 2,756,267 | 29.02 | 60.03 | 0.0010 | 0.0001 |
| Q03933 | Heat shock factor protein 2 | 60 | 1,828,433 | 1,462,167 | 2,063,067 | 0.80 | 1.13 | 0.2457 | 0.5166 |
| Q92598 | Heat shock protein 105 kDa | 97 | 438,553 | 5,339,067 | 3,570,400 | 12.17 | 8.14 | 0.0008 | 0.0016 |
| P04792 | Heat shock protein beta-1 | 23 | 40 | 234,067 | 695,710 | 5851.67 | 17,392.75 | 0.0001 | 0.0008 |
| Q9UJY1 | Heat shock protein beta-8 | 22 | 16,375 | 15,190 | 17,364 | 0.93 | 1.06 | 0.6088 | 0.7492 |
| P07900 | Heat shock protein HSP 90-alpha | 85 | 86,179 | 2,883,267 | 2,937,467 | 33.46 | 34.09 | 0.0029 | 0.0006 |
| P08238 | Heat shock protein HSP 90-beta | 83 | 145,503 | 4,792,967 | 2,217,933 | 32.94 | 15.24 | 0.0003 | 0.0043 |
| P09429 | High mobility group protein B1 | 25 | 134,723 | 260,000 | 187,963 | 1.93 | 1.40 | 0.0120 | 0.0820 |
| P26583 | High mobility group protein B2 | 24 | 19,466 | 2,220,067 | 5,347,000 | 114.05 | 274.69 | 0.0026 | 0.0010 |
| A0A3B3IU71 | Histone acetyltransferase KAT6B (Fragment) | 1 | 3951 | 58,169 | 2797 | 14.72 | 0.71 | 0.0001 | 0.0966 |
| Q92769 | Histone deacetylase 2 | 55 | 4963 | 171,967 | 4926 | 34.65 | 0.99 | 0.0001 | 0.9808 |
| P30044 | Peroxiredoxin-5, mitochondrial | 22 | 4069 | 258,147 | 456,563 | 63.44 | 112.20 | 0.0011 | 0.0030 |
| P30041 | Peroxiredoxin-6 | 25 | 51,694 | 1,386,367 | 997,103 | 26.82 | 19.29 | 0.0008 | 0.0014 |
| D3DV26 | Protein S100-A10 (Fragment) | 22 | 81,311 | 3,423,500 | 4,124,333 | 42.10 | 50.72 | 0.0063 | 0.0008 |
| P33763 | Protein S100-A5 | 11 | 16,688 | 1,242,400 | 938,803 | 74.45 | 56.26 | 0.0004 | 0.0002 |
| Q99757 | Thioredoxin, mitochondrial | 18 | 40 | 1,932,133 | 2,071,300 | 48,303.33 | 51,782.50 | 0.0048 | 0.0010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songjang, W.; Nensat, C.; Nernpermpisooth, N.; Seenak, P.; Pankhong, P.; Jumroon, N.; Kumphune, S.; Jiraviriyakul, A. Tumor-Promoting Activity and Proteomic Profiling of Cisplatin/Oxaliplatin-Derived DAMPs in Cholangiocarcinoma Cells. Int. J. Mol. Sci. 2022, 23, 10540. https://doi.org/10.3390/ijms231810540
Songjang W, Nensat C, Nernpermpisooth N, Seenak P, Pankhong P, Jumroon N, Kumphune S, Jiraviriyakul A. Tumor-Promoting Activity and Proteomic Profiling of Cisplatin/Oxaliplatin-Derived DAMPs in Cholangiocarcinoma Cells. International Journal of Molecular Sciences. 2022; 23(18):10540. https://doi.org/10.3390/ijms231810540
Chicago/Turabian StyleSongjang, Worawat, Chatchai Nensat, Nitirut Nernpermpisooth, Porrnthanate Seenak, Panyupa Pankhong, Noppadon Jumroon, Sarawut Kumphune, and Arunya Jiraviriyakul. 2022. "Tumor-Promoting Activity and Proteomic Profiling of Cisplatin/Oxaliplatin-Derived DAMPs in Cholangiocarcinoma Cells" International Journal of Molecular Sciences 23, no. 18: 10540. https://doi.org/10.3390/ijms231810540