Patients’ Stem Cells Differentiation in a 3D Environment as a Promising Experimental Tool for the Study of Amyotrophic Lateral Sclerosis
Abstract
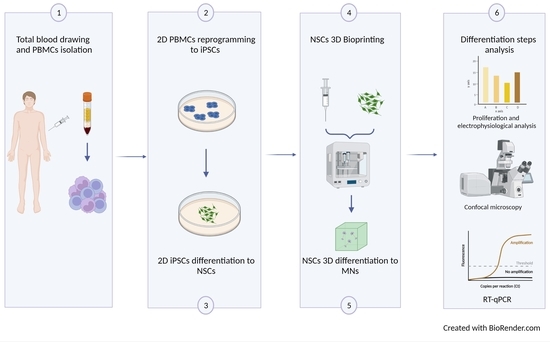
:1. Introduction
2. Results
2.1. Hydrogel Characterization Confirmed Its Optimal Features for Cells Cultures
2.2. Neural Stem Cells (NSCs) Showed Good Viability during the 3D Differentiation Process
2.3. NSCs Showed Good Differentiation When Printed in 3D
2.4. Printed Cells Expressed the Typical Markers of the Differentiation Stages
2.5. Printed Cells Maintain Their Electrophysiological Characteristics
3. Discussion
4. Materials and Methods
4.1. Enrolment of ALS Patients and Healthy Volunteers
4.2. Isolation of PBMCs
4.3. PBMCs Reprogramming
4.4. iPSC Differentiation in NSCs
4.5. Characterization of the Hydrogel
4.5.1. Moisture
4.5.2. Swelling Test
4.5.3. Porosity
4.6. Printing of NSC Cell Line
4.7. Differentiation of NSCs
4.8. Viability Assay
4.9. Immunofluorescence Analysis
4.10. RT-qPCR
4.11. Electrophysiological Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erkkinen, M.G.; Kim, M.O.; Geschwind, M.D. Clinical Neurology and Epidemiology of the Major Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2018, 10, 33118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.Y.; Zhou, Z.R.; Che, C.H.; Liu, C.Y.; He, R.L.; Huang, H.P. Genetic epidemiology of amyotrophic lateral sclerosis: A systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2017, 88, 540–549. [Google Scholar] [CrossRef]
- Bordoni, M.; Pansarasa, O.; Dell’Orco, M.; Crippa, V.; Gagliardi, S.; Sproviero, D.; Bernuzzi, S.; Diamanti, L.; Ceroni, M.; Tedeschi, G.; et al. Nuclear Phospho-SOD1 Protects DNA from Oxidative Stress Damage in Amyotrophic Lateral Sclerosis. J. Clin. Med. 2019, 8, 729. [Google Scholar] [CrossRef] [Green Version]
- Mejzini, R.; Flynn, L.L.; Pitout, I.L.; Fletcher, S.; Wilton, S.D.; Akkari, P.A. ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now? Front. Neurosci. 2019, 13, 1310. [Google Scholar] [CrossRef] [Green Version]
- Gois, A.M.; Mendonça, D.M.F.; Freire, M.A.M.; Santos, J.R. In vitro and in vivo models of amyotrophic lateral sclerosis: An updated overview. Brain Res. Bull. 2020, 159, 32–43. [Google Scholar] [CrossRef]
- Van Damme, P.; Robberecht, W.; Van Den Bosch, L. Modelling amyotrophic lateral sclerosis: Progress and possibilities. Dis. Model. Mech. 2017, 10, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Lutz, C. Mouse models of ALS: Past, present and future. Brain Res. 2018, 1693, 1–10. [Google Scholar] [CrossRef]
- Philips, T.; Rothstein, J.D. Rodent Models of Amyotrophic Lateral Sclerosis. Curr. Protoc. Pharmacol. 2015, 69, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Gregorio, S.E.; Duennwald, M.L. ALS Yeast Models-Past Success Stories and New Opportunities. Front. Mol. Neurosci. 2018, 11, 394. [Google Scholar] [CrossRef] [PubMed]
- Hawrot, J.; Imhof, S.; Wainger, B.J. Modeling cell-autonomous motor neuron phenotypes in ALS using iPSCs. Neurobiol. Dis. 2020, 134, 104680. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol. Rev. 2019, 99, 79–114. [Google Scholar] [CrossRef] [PubMed]
- Young, W.; D’Souza, S.L.; Lemischka, I.R.; Schaniel, C. Patient-specific Induced Pluripotent Stem Cells as a Platform for Disease Modeling, Drug Discovery and Precision Personalized Medicine. J. Stem Cell Res. Ther. 2012, S10, 10. [Google Scholar] [CrossRef] [Green Version]
- Guillemot, F.; Mironov, V.; Nakamura, M. Bioprinting is coming of age: Report from the International Conference on Bioprinting and Biofabrication in Bordeaux (3B’09). Biofabrication 2010, 2, 10201. [Google Scholar] [CrossRef]
- Fantini, V.; Bordoni, M.; Scocozza, F.; Conti, M.; Scarian, E.; Carelli, S.; Di Giulio, A.M.; Marconi, S.; Pansarasa, O.; Auricchio, F.; et al. Bioink Composition and Printing Parameters for 3D Modeling Neural Tissue. Cells. 2019, 8, 830. [Google Scholar] [CrossRef] [Green Version]
- Bordoni, M.; Karabulut, E.; Kuzmenko, V.; Fantini, V.; Pansarasa, O.; Cereda, C.; Gatenholm, P. 3D Printed Conductive Nanocellulose Scaffolds for the Differentiation of Human Neuroblastoma Cells. Cells. 2020, 9, 682. [Google Scholar] [CrossRef] [Green Version]
- Bordoni, M.; Scarian, E.; Rey, F.; Gagliardi, S.; Carelli, S.; Pansarasa, O.; Cereda, C. Biomaterials in Neurodegenerative Disorders: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2020, 21, 3243. [Google Scholar] [CrossRef]
- Rey, F.; Barzaghini, B.; Nardini, A.; Bordoni, M.; Zuccotti, G.V.; Cereda, C.; Raimondi, M.T.; Carelli, S. Advances in Tissue Engineering and Innovative Fabrication Techniques for 3-D-Structures: Translational Applications in Neurodegenerative Diseases. Cells. 2020, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Kolesky, D.B.; Truby, R.L.; Gladman, A.S.; Busbee, T.A.; Homan, K.A.; Lewis, J.A. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 2014, 26, 3124–3130. [Google Scholar] [CrossRef] [PubMed]
- Catros, S.; Fricain, J.C.; Guillotin, B.; Pippenger, B.; Bareille, R.; Remy, M.; Lebraud, E.; Desbat, B.; Amédée, J.; Guillemot, F. Laser-assisted bioprinting for creating on-demand patterns of human osteoprogenitor cells and nano-hydroxyapatite. Biofabrication 2011, 3, 25001. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Binder, K.W.; Albanna, M.Z.; Dice, D.; Zhao, W.; Yoo, J.J.; Atala, A. Hybrid printing of mechanically and biologically improved constructs for cartilage tissue engineering applications. Biofabrication 2013, 5, 15001. [Google Scholar] [CrossRef]
- Shevach, M.; Soffer-Tsur, N.; Fleischer, S.; Shapira, A.; Dvir, T. Fabrication of omentum-based matrix for engineering vascularized cardiac tissues. Biofabrication 2014, 6, 024101. [Google Scholar] [CrossRef]
- Vunjak Novakovic, G.; Eschenhagen, T.; Mummery, C. Myocardial tissue engineering: In vitro models. Cold Spring Harb. Perspect. Med. 2014, 4, a014076. [Google Scholar] [CrossRef] [Green Version]
- Owens, C.M.; Marga, F.; Forgacs, G.; Heesch, C.M. Biofabrication and testing of a fully cellular nerve graft. Biofabrication 2013, 5, 45007. [Google Scholar] [CrossRef] [Green Version]
- Lorber, B.; Hsiao, W.K.; Hutchings, I.M.; Martin, K.R. Adult rat retinal ganglion cells and glia can be printed by piezoelectric inkjet printing. Biofabrication 2014, 6, 015001. [Google Scholar] [CrossRef]
- Lozano, R.; Stevens, L.; Thompson, B.C.; Gilmore, K.J.; Gorkin, R., III; Stewart, E.M.; Panhuis, M.; Romero-Ortega, M.; Wallace, G.G. 3D printing of layered brain-like structures using peptide modified gellan gum substrates. Biomaterials 2015, 67, 264–273. [Google Scholar] [CrossRef]
- Maresova, P.; Hruska, J.; Klimova, B.; Barakovic, S.; Krejcar, O. Activities of Daily Living and Associated Costs in the Most Widespread Neurodegenerative Diseases: A Systematic Review. Clin. Interv. Aging. 2020, 15, 1841–1862. [Google Scholar] [CrossRef]
- Gun’ko, V.M.; Savina, I.N.; Mikhalovsky, S.V. Properties of Water Bound in Hydrogels. Gels 2017, 3, 37. [Google Scholar] [CrossRef] [PubMed]
- Shawan, M.M.A.K.; Islam, N.; Aziz, S.; Khatun, N.; Sarker, S.R.; Hossain, M.; Hossan, T.; Morshed, M.; Sarkar, M.; Shakil, S.; et al. Fabrication of Xanthan gum: Gelatin (Xnt:Gel) Hybrid Composite Hydrogels for Evaluating Skin Wound Healing Efficacy. Mod. Appl. Sci. 2019, 13, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Piola, B.; Sabbatini, M.; Gino, S.; Invernizzi, M.; Renò, F. 3D Bioprinting of Gelatin-Xanthan Gum Composite Hydrogels for Growth of Human Skin Cells. Int. J. Mol. Sci. 2022, 23, 539. [Google Scholar] [CrossRef]
- Jonoobi, M.; Harun, J.; Mathew, A.P.; Hussein, M.Z.B.; Oksman, K. Preparation of cellulose nanofibers with hydrophobic surface characteristics. Cellulose 2010, 17, 299–307. [Google Scholar] [CrossRef]
- Maiti, S.; Kumari, L. Smart Nanopolysaccharides for the Delivery of Bioactives. In Nanoarchitectonics for Smart Delivery and Drug Targeting; Alina Maria Holban, A.M.G., Ed.; William Andrew Publishing: Norwich, NY, USA, 2016; Chapter 3; pp. 67–94. [Google Scholar]
- Asohan, A.W.; Hashim, R.; Ku Ishak, K.M.; Abdul Hamid, Z.A.; Jasme, N.; Bustami, Y. Preparation and Characterisation of Cellulose Nanocrystal/Alginate/Polyethylene Glycol Diacrylate (CNC/Alg/PEGDA) Hydrogel Using Double Network Crosslinking Technique for Bioprinting Application. Appl. Sci. 2022, 12, 771. [Google Scholar] [CrossRef]
- Gu, Q.; Tomaskovic-Crook, E.; Lozano, R.; Chen, Y.; Kapsa, R.M.; Zhou, Q.; Wallace, G.G.; Crook, J.M. Functional 3D Neural Mini-Tissues from Printed Gel-Based Bioink and Human Neural Stem Cells. Adv. Healthc. Mater. 2016, 5, 1429–1438. [Google Scholar] [CrossRef] [Green Version]
- Antill-O’Brien, N.; Bourke, J.; O’Connell, C.D. Layer-By-Layer: The Case for 3D Bioprinting Neurons to Create Patient-Specific Epilepsy Models. Materials 2019, 12, 3218. [Google Scholar] [CrossRef]
- Jeon, H.; Kang, K.; Park, S.A.; Kim, W.D.; Paik, S.S.; Lee, S.H.; Jeong, J.; Choi, D. Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique. Gut Liver 2017, 11, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Bernal, A.; Arranz, L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell Mol. Life Sci. 2018, 75, 2177–2195. [Google Scholar] [CrossRef] [Green Version]
- Fong, H.; Hohenstein, K.A.; Donovan, P.J. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 2008, 26, 1931–1938. [Google Scholar] [CrossRef]
- Gao, J.; Wang, J.; Wang, Y.; Dai, W.; Lu, L. Regulation of Pax6 by CTCF during induction of mouse ES cell differentiation. PLoS ONE 2011, 6, e20954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Bohlen Und Halbach, O. Immunohistological markers for staging neurogenesis in adult hippocampus. Cell Tissue Res. 2007, 329, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Marei, H.E.S.; El-Gamal, A.; Althani, A.; Afifi, N.; Abd-Elmaksoud, A.; Farag, A.; Cenciarelli, C.; Thomas, C.; Anwarul, H. Cholinergic and dopaminergic neuronal differentiation of human adipose tissue derived mesenchymal stem cells. J. Cell Physiol. 2018, 233, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Halpain, S.; Dehmelt, L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006, 7, 224. [Google Scholar] [CrossRef]
- Cashman, N.R.; Durham, H.D.; Blusztajn, J.K.; Oda, K.; Tabira, T.; Shaw, I.T.; Dahrouge, S.; Antel, J.P. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 1992, 194, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.C.; Brey, E.M.; Pérez-Luna, V.H. A study of the intrinsic autofluorescence of poly (ethylene glycol)-co-(L-lactic acid) diacrylate. J. Fluoresc. 2012, 22, 907–913. [Google Scholar] [CrossRef]
- Xu, H.; Tan, Y.; Wang, D.; Wang, X.; An, W.; Xu, P.; Xu, S.; Wang, Y. Autofluorescence of hydrogels without a fluorophore. Soft Matter. 2019, 15, 3588–3594. [Google Scholar] [CrossRef]
- Filosto, M.; Piccinelli, S.C.; Palmieri, I.; Necchini, N.; Valente, M.; Zanella, I.; Biasiotto, G.; Lorenzo, D.D.; Cereda, C.; Padovani, A. A Novel Mutation in the Stalk Domain of KIF5A Causes a Slowly Progressive Atypical Motor Syndrome. J. Clin. Med. 2018, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Chiang, M.C.; Nicol, C.J.B.; Lin, C.H.; Chen, S.J.; Yen, C.; Huang, R.N. Nanogold induces anti-inflammation against oxidative stress induced in human neural stem cells exposed to amyloid-beta peptide. Neurochem. Int. 2021, 145, 104992. [Google Scholar] [CrossRef]
- Yi, H.; Xie, B.; Liu, B.; Wang, X.; Xu, L.; Liu, J.; Li, M.; Zhong, X.; Peng, F. Derivation and Identification of Motor Neurons from Human Urine-Derived Induced Pluripotent Stem Cells. Stem Cells Int. 2018, 2018, 3628578. [Google Scholar] [CrossRef]
- Gantenbein, B.; Croft, A.S.; Larraillet, M. Mammalian Cell Viability Methods in 3D Scaffolds for Tissue Engineering. In Fluorescence Methods for Investigation of Living Cells and Microorganisms; Grigoryeva, N., Ed.; Intech Open: London, UK, 2020; pp. 1–25. [Google Scholar]
- García-Couce, J.; Vernhes, M.; Bada, N.; Agüero, L.; Valdés, O.; Alvarez-Barreto, J.; Fuentes, G.; Almirall, A.; Cruz, L.J. Synthesis and Evaluation of AlgNa-g-Poly(QCL-co-HEMA) Hydrogels as Platform for Chondrocyte Proliferation and Controlled Release of Betamethasone. Int. J. Mol. Sci. 2021, 22, 5730. [Google Scholar] [CrossRef] [PubMed]
- Weitkamp, J.T.; Wöltje, M.; Nußpickel, B.; Schmidt, F.N.; Aibibu, D.; Bayer, A.; Eglin, D.; Armiento, A.R.; Arnold, P.; Cherif, C.; et al. Silk Fiber-Reinforced Hyaluronic Acid-Based Hydrogel for Cartilage Tissue Engineering. Int. J. Mol. Sci. 2021, 22, 3635. [Google Scholar] [CrossRef] [PubMed]
- Leu Alexa, R.; Cucuruz, A.; Ghițulică, C.-D.; Voicu, G.; Stamat, L.-R.; Dinescu, S.; Vlasceanu, G.M.; Stavarache, C.; Ianchis, R.; Iovu, H.; et al. 3D Printable Composite Biomaterials Based on GelMA and Hydroxyapatite Powders Doped with Cerium Ions for Bone Tissue Regeneration. Int. J. Mol. Sci. 2022, 23, 1841. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zeng, X.; Hu, J.; Chen, Q. Characterization of Nestin, a Selective Marker for Bone Marrow Derived Mesenchymal Stem Cells. Stem Cells Int. 2015, 2015, 762098. [Google Scholar] [CrossRef] [Green Version]
- Pagin, M.; Pernebrink, M.; Giubbolini, S.; Barone, C.; Sambruni, G.; Zhu, Y.; Chiara, M.; Ottolenghi, S.; Pavesi, G.; Wei, C.L.; et al. Sox2 controls neural stem cell self-renewal through a Fos-centered gene regulatory network. Stem Cells. 2021, 39, 1107–1119. [Google Scholar] [CrossRef]
- Archer, T.C.; Jin, J.; Casey, E.S. Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Dev. Biol. 2011, 350, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Elkouris, M.; Balaskas, N.; Poulou, M.; Politis, P.K.; Panayiotou, E.; Malas, S.; Thomaidou, D.; Remboutsika, E. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells 2011, 29, 89–98. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, C.T.; Chen, J.; Pankratz, M.T.; Xi, J.; Li, J.; Yang, Y.; Lavaute, T.M.; Li, X.J.; Ayala, M.; et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 2010, 7, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Xue, H.; Mattson, M.P.; Rao, M.S. Stage-dependent Olig2 expression in motor neurons and oligodendrocytes differentiated from embryonic stem cells. Stem Cells Dev. 2007, 16, 131–141. [Google Scholar] [CrossRef]
- Tischfield, M.A.; Baris, H.N.; Wu, C.; Rudolph, G.; Van Maldergem, L.; He, W.; Chan, W.M.; Andrews, C.; Demer, J.L.; Robertson, R.L.; et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 2010, 140, 74–87. [Google Scholar] [CrossRef] [Green Version]
- Veit, W. Choline Acetyltransferase (xPharm: The Comprehensive Pharmacology Reference); Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar] [CrossRef]
- Measuring Cell Fluorescence Using ImageJ. Available online: http://theolb.readthedocs.io/en/latest/imaging/measuring-cell-fluorescence-using-imagej.html#measuring-cell-fluorescence-using-imagej (accessed on 13 January 2021).
- Kenan, S.; Liang, H.; Goodman, H.J.; Jacobs, A.J.; Chan, A.; Grande, D.A.; Levin, A.S. 5-Aminolevulinic acid tumor paint and photodynamic therapy for myxofibrosarcoma: An in vitro study. J. Orthop. Surg. Res. 2020, 15, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Li, P.; Wang, M.; Zhang, J.; Wang, W. Imaging the brain in 3D using a combination of CUBIC and immunofluorescence staining. Biomed. Opt Express. 2019, 10, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.A. Brief Introduction to the Transduction of Neural Activity into Fos Signal. Dev. Reprod. 2015, 19, 61–67. [Google Scholar] [CrossRef] [PubMed]






| Primary Antibody | Secondary Antibody | Brand |
|---|---|---|
| Nestin 1:100 | CFTM 594 goat anti-mouse 1:500 | Abcam (Cambridge, UK) |
| SOX2 1:250 | CFTM 488A goat anti-rabbit 1:500 | Proteintech (Chicago, IL, USA) |
| SOX1 1:200 | CFTM 488A goat anti-rabbit 1:400 | Abcam (Cambridge, UK) |
| PAX6 1:100 | CFTM 594 goat anti-mouse 1:400 | Invitrogen (Carlsbad, CA, USA) |
| Olig2 1:400 | CFTM 488A goat anti-rabbit 1:500 | Novus Biologicals (Centennial, CO, USA) |
| TUBB3 1:400 | CFTM 488A goat anti-rabbit 1:500 | Abcam (Cambridge, UK) |
| Chat 1:200 | CFTM 594 goat anti-mouse 1:400 | Novus Biologicals (Centennial, CO, USA) |
| Gene | Primer Sequences (5′-3′) | Annealing Temperature |
|---|---|---|
| Nestin | F GGA AGA GAA CCT GGG AAA GG R GAT TCA GCT CTG CCT CAT CC | 60 °C |
| SOX2 | F AGTCTCCAAGCGACGAAAAA R TTTCACGTTTGCAACTGTCC | 60 °C |
| SOX1 | F AAATACTGGAGACGAACGCC R AACCCAAGTCTGGTGTCAGC | 60 °C |
| PAX6 | F TGTGTGCTCTGAAGGTCAGG R CTGGAGCTCTGTTTGGAAGG | 60 °C |
| TUBB3 | F CAGATGTTCGATGCCAAGAA R GGGATCCACTCCACGAAGTA | 60 °C |
| MAP2 | F AGGGCTGGTAGGTTGGATCT R TGTGTCTCTGCCTTTGTATC | 60 °C |
| GAPDH | F CAG CAA GAG CAC AAG AGG AAG R CAA CTG TGA GGA GGG GAG ATT | 60 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scarian, E.; Bordoni, M.; Fantini, V.; Jacchetti, E.; Raimondi, M.T.; Diamanti, L.; Carelli, S.; Cereda, C.; Pansarasa, O. Patients’ Stem Cells Differentiation in a 3D Environment as a Promising Experimental Tool for the Study of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2022, 23, 5344. https://doi.org/10.3390/ijms23105344
Scarian E, Bordoni M, Fantini V, Jacchetti E, Raimondi MT, Diamanti L, Carelli S, Cereda C, Pansarasa O. Patients’ Stem Cells Differentiation in a 3D Environment as a Promising Experimental Tool for the Study of Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences. 2022; 23(10):5344. https://doi.org/10.3390/ijms23105344
Chicago/Turabian StyleScarian, Eveljn, Matteo Bordoni, Valentina Fantini, Emanuela Jacchetti, Manuela Teresa Raimondi, Luca Diamanti, Stephana Carelli, Cristina Cereda, and Orietta Pansarasa. 2022. "Patients’ Stem Cells Differentiation in a 3D Environment as a Promising Experimental Tool for the Study of Amyotrophic Lateral Sclerosis" International Journal of Molecular Sciences 23, no. 10: 5344. https://doi.org/10.3390/ijms23105344