Redox-Active Metal Ions and Amyloid-Degrading Enzymes in Alzheimer’s Disease
Abstract
:1. Introduction
2. Cu(I/II)
2.1. Cu(I/II) Distributions in the Nervous System
2.2. Regulations of Cu by Metallochaperones
2.2.1. Copper Chaperone for Superoxide Dismutase (CCS)
2.2.2. Antioxidant Protein 1 (Atox1)
2.2.3. Cytochrome C Oxidase Assembly: Cox11, Cox17, Cox19, Sco1, and Sco2
2.3. Uptake of Cu(I/II) through Blood–Brain Barrier
2.3.1. Copper Transporter-1 (Ctr1), Ctr2, and Ctr6
2.3.2. ATP7A and ATP7B
2.3.3. Glutathione (GSH)
2.3.4. Metallothioneins (MTs)
2.4. Cu in Normal and Diseased Conditions
2.4.1. Cu in Nervous Systems under Normal Conditions
2.4.2. Cu under Diseased Conditions
2.5. Regulators of Cu(I/II)
3. Fe(II/III)
3.1. Fe(II/III) Distributions in the Nervous Systems
3.2. Homeostasis of Fe
3.2.1. Fe(II/III) Cross the BBB by Involvement of Tf
3.2.2. Fe(II/III) Transport without Tf
3.3. Physiological and Pathological Functions of Fe in Nervous Systems
3.3.1. Fe under Normal Conditions
3.3.2. Fe in Diseased Conditions
3.4. Regulators of Fe(II/III)
4. Amyloid Degrading Enzymes (ADE)
4.1. Neprilysin (NEP)
4.2. Insulin-Degrading Enzyme (IDE)
4.3. ADAM10
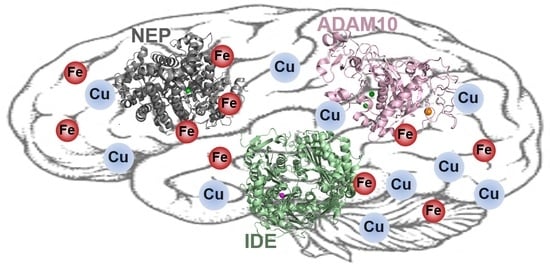
5. Redox-Active Metal Ions with ADE
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Savelieff, M.G.; Nam, G.; Kang, J.; Lee, H.J.; Lee, M.; Lim, M.H. Development of Multifunctional Molecules as Potential Therapeutic Candidates for Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the Last Decade. Chem. Rev. 2019, 119, 1221–1322. [Google Scholar] [CrossRef] [PubMed]
- Gromadzka, G.; Tarnacka, B.; Flaga, A.; Adamczyk, A. Copper Dyshomeostasis in Neurodegenerative Diseases-Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 9259. [Google Scholar] [CrossRef] [PubMed]
- Barnham, K.J.; Bush, A.I. Biological metals and metal-targeting compounds in major neurodegenerative diseases. Chem. Soc. Rev. 2014, 43, 6727–6749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kepp, K.P. Bioinorganic chemistry of Alzheimer’s disease. Chem. Rev. 2012, 112, 5193–5239. [Google Scholar] [CrossRef] [Green Version]
- Bonda, D.J.; Lee, H.G.; Blair, J.A.; Zhu, X.; Perry, G.; Smith, M.A. Role of metal dyshomeostasis in Alzheimer’s disease. Metallomics 2011, 3, 267–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolognin, S.; Messori, L.; Zatta, P. Metal ion physiopathology in neurodegenerative disorders. Neuromol. Med. 2009, 11, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef]
- Millecamps, S.; Julien, J.P. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 2013, 14, 161–176. [Google Scholar] [CrossRef]
- Maday, S.; Twelvetrees, A.E.; Moughamian, A.J.; Holzbaur, E.L. Axonal transport: Cargo-specific mechanisms of motility and regulation. Neuron 2014, 84, 292–309. [Google Scholar] [CrossRef] [Green Version]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef]
- De Ricco, R.; Valensin, D.; Dell’Acqua, S.; Casella, L.; Hureau, C.; Faller, P. Copper(I/II), a/b-Synuclein and Amyloid-b: Menage a Trois? ChemBioChem 2015, 16, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Faller, P.; Hureau, C.; Berthoumieu, O. Role of metal ions in the self-assembly of the Alzheimer’s amyloid-b peptide. Inorg. Chem. 2013, 52, 12193–12206. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. The amyloid beta peptide: A chemist’s perspective. Role in Alzheimer’s and fibrillization. Chem. Rev. 2012, 112, 5147–5192. [Google Scholar] [CrossRef] [PubMed]
- Squitti, R.; Faller, P.; Hureau, C.; Granzotto, A.; White, A.R.; Kepp, K.P. Copper imbalance in Alzheimer’s disease and its link with the amyloid hypothesis: Towards a combined clinical, chemical, and genetic etiology. J. Alzheimers Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Nalivaeva, N.N.; Turner, A.J. Targeting amyloid clearance in Alzheimer’s disease as a therapeutic strategy. Br. J. Pharmacol. 2019, 176, 3447–3463. [Google Scholar] [CrossRef] [PubMed]
- Ries, M.; Sastre, M. Mechanisms of Ab clearance and degradation by glial cells. Front. Aging Neurosci. 2016, 8, 160. [Google Scholar] [CrossRef] [Green Version]
- Nalivaeva, N.N.; Turner, A.J. Role of ageing and oxidative stress in regulation of amyloid-degrading enzymes and development of neurodegeneration. Curr. Aging Sci. 2017, 10, 32–40. [Google Scholar] [CrossRef]
- Mawuenyega, K.G.; Sigurdson, W.; Ovod, V.; Munsell, L.; Kasten, T.; Morris, J.C.; Yarasheski, K.E.; Bateman, R.J. Decreased clearance of CNS b-amyloid in Alzheimer’s disease. Science 2010, 330, 1774. [Google Scholar] [CrossRef] [Green Version]
- Hemming, M.L.; Patterson, M.; Reske-Nielsen, C.; Lin, L.; Isacson, O.; Selkoe, D.J. Reducing amyloid plaque burden via ex vivo gene delivery of an Ab-degrading protease: A novel therapeutic approach to Alzheimer disease. PLoS Med. 2007, 4, e262. [Google Scholar] [CrossRef] [Green Version]
- Gaggelli, E.; Kozlowski, H.; Valensin, D.; Valensin, G. Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis). Chem. Rev. 2006, 106, 1995–2044. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, T.V.; Sorokin, D.Y.; Hagen, W.R.; Khrenova, M.G.; Muyzer, G.; Rakitina, T.V.; Shabalin, I.G.; Trofimov, A.A.; Tsallagov, S.I.; Popov, V.O. Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase. Proc. Natl. Acad. Sci. USA 2020, 117, 5280–5290. [Google Scholar] [CrossRef] [PubMed]
- Greenough, M.A.; Camakaris, J.; Bush, A.I. Metal dyshomeostasis and oxidative stress in Alzheimer’s disease. Neurochem. Int. 2013, 62, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.T.; Anderson, C.M. Cardiac cytochrome C oxidase activity and contents of subunits 1 and 4 are altered in offspring by low prenatal copper intake by rat dams. J. Nutr. 2008, 138, 1269–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, K.T.; Prohaska, J.R. Copper deficiency in rodents alters dopamine b-mono-oxygenase activity, mRNA and protein level. Br. J. Nutr. 2009, 102, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Bisaglia, M.; Bubacco, L. Copper Ions and Parkinson’s Disease: Why Is Homeostasis So Relevant? Biomolecules 2020, 10, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jett, K.A.; Leary, S.C. Building the CuA site of cytochrome c oxidase: A complicated, redox-dependent process driven by a surprisingly large complement of accessory proteins. J. Biol. Chem. 2018, 293, 4644–4652. [Google Scholar] [CrossRef] [Green Version]
- Kozma, M.; Szerdahelyi, P.; Kasa, P. Histochemical detection of zinc and copper in various neurons of the central nervous system. Acta Histochem. 1981, 69, 12–17. [Google Scholar] [CrossRef]
- Stockel, J.; Safar, J.; Wallace, A.C.; Cohen, F.E.; Prusiner, S.B. Prion protein selectively binds copper(II) ions. Biochemistry 1998, 37, 7185–7193. [Google Scholar] [CrossRef] [PubMed]
- Kardos, J.; Heja, L.; Simon, A.; Jablonkai, I.; Kovacs, R.; Jemnitz, K. Copper signalling: Causes and consequences. Cell Commun. Signal. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.R.; Qin, K.; Herms, J.W.; Madlung, A.; Manson, J.; Strome, R.; Fraser, P.E.; Kruck, T.; von Bohlen, A.; Schulz-Schaeffer, W.; et al. The cellular prion protein binds copper in vivo. Nature 1997, 390, 684–687. [Google Scholar] [CrossRef]
- Bush, A.I. Metals and neuroscience. Curr. Opin. Chem. Biol. 2000, 4, 184–191. [Google Scholar] [CrossRef]
- Atwood, C.S.; Huang, X.; Moir, R.D.; Tanzi, R.E.; Bush, A.I. Role of free radicals and metal ions in the pathogenesis of Alzheimer’s disease. Met. Ions Biol. Syst. 1999, 36, 309–364. [Google Scholar] [PubMed]
- Schlief, M.L.; Gitlin, J.D. Copper homeostasis in the CNS: A novel link between the NMDA receptor and copper homeostasis in the hippocampus. Mol. Neurobiol. 2006, 33, 81–90. [Google Scholar] [CrossRef]
- Kardos, J.; Kovacs, I.; Hajos, F.; Kalman, M.; Simonyi, M. Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability. Neurosci. Lett. 1989, 103, 139–144. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevitt, T.; Ohrvik, H.; Thiele, D.J. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 2012, 1823, 1580–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Wang, L.; Li, D.; Zhao, C.; Li, J.; Liu, T. Exploring the extended biological functions of the human copper chaperone of superoxide dismutase 1. Protein J. 2019, 38, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Skopp, A.; Boyd, S.D.; Ullrich, M.S.; Liu, L.; Winkler, D.D. Copper-zinc superoxide dismutase (Sod1) activation terminates interaction between its copper chaperone (Ccs) and the cytosolic metal-binding domain of the copper importer Ctr1. Biometals 2019, 32, 695–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klomp, L.W.; Lin, S.J.; Yuan, D.S.; Klausner, R.D.; Culotta, V.C.; Gitlin, J.D. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J. Biol. Chem. 1997, 272, 9221–9226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Annu. Rev. Biochem. 2010, 79, 537–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolgova, N.V.; Yu, C.; Cvitkovic, J.P.; Hodak, M.; Nienaber, K.H.; Summers, K.L.; Cotelesage, J.J.H.; Bernholc, J.; Kaminski, G.A.; Pickering, I.J.; et al. Binding of copper and cisplatin to Atox1 is mediated by glutathione through the formation of metal-sulfur clusters. Biochemistry 2017, 56, 3129–3141. [Google Scholar] [CrossRef] [PubMed]
- Hatori, Y.; Lutsenko, S. The Role of Copper Chaperone Atox1 in Coupling Redox Homeostasis to Intracellular Copper Distribution. Antioxidants 2016, 5, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatori, Y.; Inouye, S.; Akagi, R. Thiol-based copper handling by the copper chaperone Atox1. IUBMB Life 2017, 69, 246–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blockhuys, S.; Zhang, X.; Wittung-Stafshede, P. Single-cell tracking demonstrates copper chaperone Atox1 to be required for breast cancer cell migration. Proc. Natl. Acad. Sci. USA 2020, 117, 2014–2019. [Google Scholar] [CrossRef] [Green Version]
- Carr, H.S.; Maxfield, A.B.; Horng, Y.C.; Winge, D.R. Functional analysis of the domains in Cox11. J. Biol. Chem. 2005, 280, 22664–22669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, H.S.; George, G.N.; Winge, D.R. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002, 277, 31237–31242. [Google Scholar] [CrossRef] [Green Version]
- Radin, I.; Gey, U.; Kost, L.; Steinebrunner, I.; Rödel, G. The mitochondrial copper chaperone COX11 plays an auxiliary role in the defence against oxidative stress. BioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Maxfield, A.B.; Heaton, D.N.; Winge, D.R. Cox17 is functional when tethered to the mitochondrial inner membrane. J. Biol. Chem. 2004, 279, 5072–5080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inesi, G. Molecular features of copper binding proteins involved in copper homeostasis. IUBMB Life 2017, 69, 211–217. [Google Scholar] [CrossRef]
- Heaton, D.N.; George, G.N.; Garrison, G.; Winge, D.R. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 2001, 40, 743–751. [Google Scholar] [CrossRef]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20531–20535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigby, K.; Zhang, L.; Cobine, P.A.; George, G.N.; Winge, D.R. characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 10233–10242. [Google Scholar] [PubMed] [Green Version]
- Garcia, L.; Mansilla, N.; Ocampos, N.; Pagani, M.A.; Welchen, E.; Gonzalez, D.H. The mitochondrial copper chaperone COX19 influences copper and iron homeostasis in arabidopsis. Plant Mol. Biol. 2019, 99, 621–638. [Google Scholar] [CrossRef]
- Petruzzella, V.; Tiranti, V.; Fernandez, P.; Ianna, P.; Carrozzo, R.; Zeviani, M. Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 1998, 54, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Horng, Y.C.; Leary, S.C.; Cobine, P.A.; Young, F.B.; George, G.N.; Shoubridge, E.A.; Winge, D.R. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 2005, 280, 34113–34122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekim Kocabey, A.; Kost, L.; Gehlhar, M.; Rodel, G.; Gey, U. Mitochondrial Sco proteins are involved in oxidative stress defense. Redox Biol. 2019, 21, 101079. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.C.; Sue, C.; Banting, G.S.; Yang, H.; Glerum, D.M.; Hendrickson, W.A.; Schon, E.A. Crystal structure of human SCO1: Implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 2005, 280, 15202–15211. [Google Scholar] [CrossRef] [Green Version]
- Balatri, E.; Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S. Solution structure of Sco1: A thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure 2003, 11, 1431–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leary, S.C.; Kaufman, B.A.; Pellecchia, G.; Guercin, G.H.; Mattman, A.; Jaksch, M.; Shoubridge, E.A. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004, 13, 1839–1848. [Google Scholar] [CrossRef] [Green Version]
- Leary, S.C.; Cobine, P.A.; Kaufman, B.A.; Guercin, G.H.; Mattman, A.; Palaty, J.; Lockitch, G.; Winge, D.R.; Rustin, P.; Horvath, R.; et al. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007, 5, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, Z.N.; Cobine, P.A.; Leary, S.C. The mitochondrion: A central architect of copper homeostasis. Metallomics 2017, 9, 1501–1512. [Google Scholar] [CrossRef]
- Leary, S.C.; Sasarman, F.; Nishimura, T.; Shoubridge, E.A. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 2009, 18, 2230–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Monnot, A.D. Regulation of brain iron and copper homeostasis by brain barrier systems: Implication in neurodegenerative diseases. Pharmacol. Ther. 2012, 133, 177–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Metabolism and functions of copper in brain. Prog. Neurobiol. 2014, 116, 33–57. [Google Scholar] [CrossRef] [PubMed]
- Mezzaroba, L.; Alfieri, D.F.; Colado Simao, A.N.; Vissoci Reiche, E.M. The role of zinc, copper, manganese and iron in neurodegenerative diseases. Neurotoxicology 2019, 74, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Rae, T.D.; Schmidt, P.J.; Pufahl, R.A.; Culotta, V.C.; O’Halloran, T.V. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science 1999, 284, 805–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassani, S.; Ghaffari, P.; Chahardouli, B.; Alimoghaddam, K.; Ghavamzadeh, A.; Alizadeh, S.; Ghaffari, S.H. Disulfiram/copper causes ROS levels alteration, cell cycle inhibition, and apoptosis in acute myeloid leukaemia cell lines with modulation in the expression of related genes. Biomed. Pharmacother. 2018, 99, 561–569. [Google Scholar] [CrossRef]
- Nishito, Y.; Kambe, T. Absorption mechanisms of iron, copper, and zinc: An overview. J. Nutr. Sci. Vitaminol. 2018, 64, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef] [Green Version]
- Hassett, R.; Kosman, D.J. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Jiang, Y.; Yang, Y.; Peng, Y.; Li, C. Copper metabolism in Saccharomyces cerevisiae: An update. Biometals 2021, 34, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Gitschier, J. hCTR1: A human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 7481–7486. [Google Scholar] [CrossRef] [Green Version]
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402. [Google Scholar] [CrossRef]
- Guo, Y.; Smith, K.; Lee, J.; Thiele, D.J.; Petris, M.J. Identification of methionine-rich clusters that regulate copper-stimulated endocytosis of the human Ctr1 copper transporter. J. Biol. Chem. 2004, 279, 17428–17433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefaniak, E.; Plonka, D.; Drew, S.C.; Bossak-Ahmad, K.; Haas, K.L.; Pushie, M.J.; Faller, P.; Wezynfeld, N.E.; Bal, W. The N-terminal 14-mer model peptide of human Ctr1 can collect Cu(ii) from albumin. Implications for copper uptake by Ctr1. Metallomics 2018, 10, 1723–1727. [Google Scholar] [CrossRef]
- Shenberger, Y.; Marciano, O.; Gottlieb, H.E.; Ruthstein, S. Insights into the N-terminal Cu(II) and Cu(I) binding sites of the human copper transporter CTR1. J. Coord. Chem. 2018, 71, 1985–2002. [Google Scholar] [CrossRef] [Green Version]
- Eisses, J.F.; Kaplan, J.H. Molecular characterization of hCTR1, the human copper uptake protein. J. Biol. Chem. 2002, 277, 29162–29171. [Google Scholar] [CrossRef] [Green Version]
- Klomp, A.E.; Juijn, J.A.; van der Gun, L.T.; van den Berg, I.E.; Berger, R.; Klomp, L.W. The N-terminus of the human copper transporter 1 (hCTR1) is localized extracellularly, and interacts with itself. Biochem. J. 2003, 370, 881–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Lutsenko, S. Human copper transporters: Mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 2009, 1, 1125–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Pena, M.M.; Nose, Y.; Thiele, D.J. Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 2002, 277, 4380–4387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klomp, A.E.; Tops, B.B.; Van Denberg, I.E.; Berger, R.; Klomp, L.W. Biochemical characterization and subcellular localization of human copper transporter 1 (hCTR1). Biochem. J. 2002, 364, 497–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Peng, J.; Zeng, F.; Zhang, K.; Liu, J.; Li, X.; Ouyang, Q.; Wang, G.; Wang, L.; Liu, Z.; et al. Association between polymorphisms in CTR1, CTR2, ATP7A, and ATP7B and platinum resistance in epithelial ovarian cancer. Int. J. Clin. Pharamacol. Ther. 2017, 55, 774–780. [Google Scholar] [CrossRef]
- van den Berghe, P.V.; Folmer, D.E.; Malingre, H.E.; van Beurden, E.; Klomp, A.E.; van de Sluis, B.; Merkx, M.; Berger, R.; Klomp, L.W. Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem. J. 2007, 407, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Bertinato, J.; Swist, E.; Plouffe, L.J.; Brooks, S.P.; L’Abbe, M.R. Ctr2 is partially localized to the plasma membrane and stimulates copper uptake in COS-7 cells. Biochem. J. 2008, 409, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Rees, E.M.; Thiele, D.J. Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 2007, 282, 21629–21638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portnoy, M.E.; Schmidt, P.J.; Rogers, R.S.; Culotta, V.C. Metal transporters that contribute copper to metallochaperones in Saccharomyces cerevisiae. Mol. Genet. Genom. Med. 2001, 265, 873–882. [Google Scholar] [CrossRef]
- Bellemare, D.R.; Shaner, L.; Morano, K.A.; Beaudoin, J.; Langlois, R.; Labbe, S. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 2002, 277, 46676–46686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plante, S.; Normant, V.; Ramos-Torres, K.M.; Labbe, S. Cell-surface copper transporters and superoxide dismutase 1 are essential for outgrowth during fungal spore germination. J. Biol. Chem. 2017, 292, 11896–11914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strausak, D.; La Fontaine, S.; Hill, J.; Firth, S.D.; Lockhart, P.J.; Mercer, J.F. The role of GMXCXXC metal binding sites in the copper-induced redistribution of the Menkes protein. J. Biol. Chem. 1999, 274, 11170–11177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephenson, S.E.; Dubach, D.; Lim, C.M.; Mercer, J.F.; La Fontaine, S. A single PDZ domain protein interacts with the Menkes copper ATPase, ATP7A. A new protein implicated in copper homeostasis. J. Biol. Chem. 2005, 280, 33270–33279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, M.J.; Jones, E.E.; Levy, E.R.; Martin, R.L.; Ponnambalam, S.; Monaco, A.P. Identification of a di-leucine motif within the C terminus domain of the Menkes disease protein that mediates endocytosis from the plasma membrane. J. Cell Sci. 1999, 112, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Dolgova, N.V.; Dmitriev, O.Y. Dynamics of the metal binding domains and regulation of the human copper transporters ATP7B and ATP7A. IUBMB life 2017, 69, 226–235. [Google Scholar] [CrossRef]
- Walker, J.M.; Huster, D.; Ralle, M.; Morgan, C.T.; Blackburn, N.J.; Lutsenko, S. The N-terminal metal-binding site 2 of the Wilson’s Disease Protein plays a key role in the transfer of copper from Atox1. J. Biol. Chem. 2004, 279, 15376–15384. [Google Scholar] [CrossRef] [Green Version]
- Achila, D.; Banci, L.; Bertini, I.; Bunce, J.; Ciofi-Baffoni, S.; Huffman, D.L. Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Natl. Acad. Sci. USA 2006, 103, 5729–5734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huster, D.; Lutsenko, S. The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson’s disease protein. J. Biol. Chem. 2003, 278, 32212–32218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, N.; Tsivkovskii, R.; Tsivkovskaia, N.; Lutsenko, S. The copper-transporting ATPases, menkes and wilson disease proteins, have distinct roles in adult and developing cerebellum. J. Biol. Chem. 2005, 280, 9640–9645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogorek, M.; Lenartowicz, M.; Starzynski, R.; Jonczy, A.; Staron, R.; Doniec, A.; Krzeptowski, W.; Bednarz, A.; Pierzchala, O.; Lipinski, P.; et al. Atp7a and Atp7b regulate copper homeostasis in developing male germ cells in mice. Metallomics 2017, 9, 1288–1303. [Google Scholar] [CrossRef] [PubMed]
- Nyasae, L.; Bustos, R.; Braiterman, L.; Eipper, B.; Hubbard, A. Dynamics of endogenous ATP7A (Menkes protein) in intestinal epithelial cells: Copper-dependent redistribution between two intracellular sites. Am. J. Physiol. Gastrointest Liver Physiol. 2007, 292, G1181–G1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pase, L.; Voskoboinik, I.; Greenough, M.; Camakaris, J. Copper stimulates trafficking of a distinct pool of the Menkes copper ATPase (ATP7A) to the plasma membrane and diverts it into a rapid recycling pool. Biochem. J. 2004, 378, 1031–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadini-Buoninsegni, F.; Smeazzetto, S. Mechanisms of charge transfer in human copper ATPases ATP7A and ATP7B. IUBMB Life 2017, 69, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Hellman, N.E.; Kono, S.; Mancini, G.M.; Hoogeboom, A.J.; De Jong, G.J.; Gitlin, J.D. Mechanisms of copper incorporation into human ceruloplasmin. J. Biol. Chem. 2002, 277, 46632–46638. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Adeoye, O.; Olawumi, J.; Opeyemi, A.; Christiania, O. Review on the role of glutathione on oxidative stress and infertility. JBRA Assist. Reprod. 2018, 22, 61–66. [Google Scholar] [CrossRef]
- Pastore, A.; Piemonte, F.; Locatelli, M.; Lo Russo, A.; Gaeta, L.M.; Tozzi, G.; Federici, G. Determination of blood total, reduced, and oxidized glutathione in pediatric subjects. Clin. Chem. 2001, 47, 1467–1469. [Google Scholar] [CrossRef] [Green Version]
- Halprin, K.M.; Ohkawara, A. The measurement of glutathione in human epidermis using glutathione reductase. J. Invest. Dermatol. 1967, 48, 149–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefaniak, E.; Plonka, D.; Szczerba, P.; Wezynfeld, N.E.; Bal, W. Copper Transporters? Glutathione Reactivity of Products of Cu-Abeta Digestion by Neprilysin. Inorg. Chem. 2020, 59, 4186–4190. [Google Scholar] [CrossRef] [PubMed]
- Maryon, E.B.; Molloy, S.A.; Kaplan, J.H. Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am. J. Physiol. Cell Physiol. 2013, 304, C768–C779. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, A.; Chakraborty, K.; Shukla, A. Cellular copper homeostasis: Current concepts on its interplay with glutathione homeostasis and its implication in physiology and human diseases. Metallomics 2017, 9, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.M.; Cater, M.A.; Mercer, J.F.; La Fontaine, S. Copper-dependent interaction of glutaredoxin with the N termini of the copper-ATPases (ATP7A and ATP7B) defective in Menkes and Wilson diseases. Biochem. Biophys. Res. Commun. 2006, 348, 428–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, W.C.J.; McInnes, K.T.; Cater, M.A.; Winnall, W.R.; McKirdy, R.; Yu, Y.; Taylor, P.E.; Ke, B.X.; Richardson, D.R.; Mercer, J.F.B.; et al. Role of glutaredoxin1 and glutathione in regulating the activity of the copper-transporting P-type ATPases, ATP7A and ATP7B. J. Biol. Chem. 2010, 285, 27111–27121. [Google Scholar] [CrossRef] [Green Version]
- Ziller, A.; Fraissinet-Tachet, L. Metallothionein diversity and distribution in the tree of life: A multifunctional protein. Metallomics 2018, 10, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Calvo, J.; Jung, H.; Meloni, G. Copper metallothioneins. IUBMB Life 2017, 69, 236–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.Z.; Lin, Y.J.; Zhang, Q. Metallothioneins enhance chromium detoxification through scavenging ROS and stimulating metal chelation in Oryza sativa. Chemosphere 2019, 220, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tan, S.; Wang, Y.; Luo, J.; Long, Y.; Mei, X.; Tang, Y. Role of metallothionein-1 and metallothionein-2 in the neuroprotective mechanism of sevoflurane preconditioning in mice. J. Mol. Neurosci. 2020, 70, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Vasak, M. Possible role for metallothionein in protection against radiation-induced oxidative stress. Kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Biochim. Biophys. Acta 1985, 827, 36–44. [Google Scholar] [CrossRef]
- Binz, P.-A.; Kägi, J.H.R. Metallothionein: Molecular evolution and classification. In Metallothionein IV; Klaassen, C.D., Ed.; Birkhauser: Basel, Switzerland, 1999; pp. 7–13. [Google Scholar]
- Sakatoku, A.; Ishikawa, M.; Yamazaki, K.; Nakamachi, T.; Kamachi, H.; Tanaka, D.; Nakamura, S. Molecular identification, characterization, and expression analysis of a metallothionein gene from septifer virgatus. Mar. Biothenol. 2020, 22, 488–497. [Google Scholar] [CrossRef]
- Cherian, M.G.; Jayasurya, A.; Bay, B.H. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat. Res. 2003, 533, 201–209. [Google Scholar] [CrossRef]
- Rahman, M.T.; Haque, N.; Abu Kasim, N.H.; De Ley, M. Origin, function, and fate of metallothionein in human blood. Rev. Physiol. Biochem. Pharmacol. 2017, 173, 41–62. [Google Scholar]
- Kim, B.E.; Nevitt, T.; Thiele, D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008, 4, 176–185. [Google Scholar] [CrossRef]
- Alwan, K.B.; Welch, E.F.; Blackburn, N.J. Catalytic M center of copper monooxygenases probed by rational design. effects of selenomethionine and histidine substitution on structure and reactivity. Biochemistry 2019, 58, 4436–4446. [Google Scholar] [CrossRef] [PubMed]
- Hartter, D.E.; Barnea, A. Evidence for release of copper in the brain: Depolarization-induced release of newly taken-up 67copper. Synapse 1988, 2, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Schlief, M.L.; Craig, A.M.; Gitlin, J.D. NMDA receptor activation mediates copper homeostasis in hippocampal neurons. J. Neurosci. 2005, 25, 239–246. [Google Scholar] [CrossRef] [Green Version]
- Alies, B.; Renaglia, E.; Rozga, M.; Bal, W.; Faller, P.; Hureau, C. Cu(II) affinity for the Alzheimer’s peptide: Tyrosine fluorescence studies revisited. Anal. Chem. 2013, 85, 1501–1508. [Google Scholar] [CrossRef]
- Weibull, M.G.M.; Simonsen, S.; Oksbjerg, C.R.; Tiwari, M.K.; Hemmingsen, L. Effects of Cu(II) on the aggregation of amyloid-b. J. Biol. Inorg. Chem. 2019, 24, 1197–1215. [Google Scholar] [CrossRef]
- Drew, S.C.; Noble, C.J.; Masters, C.L.; Hanson, G.R.; Barnham, K.J. Pleomorphic copper coordination by Alzheimer’s disease amyloid-beta peptide. J. Am. Chem. Soc. 2009, 131, 1195–1207. [Google Scholar] [CrossRef]
- Summers, K.L.; Schilling, K.M.; Roseman, G.; Markham, K.A.; Dolgova, N.V.; Kroll, T.; Sokaras, D.; Millhauser, G.L.; Pickering, I.J.; George, G.N. X-ray Absorption spectroscopy investigations of copper(II) coordination in the human amyloid b peptide. Inorg. Chem. 2019, 58, 6294–6311. [Google Scholar] [CrossRef] [PubMed]
- Duce, J.A.; Bush, A.I. Biological metals and Alzheimer’s disease: Implications for therapeutics and diagnostics. Prog. Neurobiol. 2010, 92, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Branch, T.; Barahona, M.; Dodson, C.A.; Ying, L. Kinetic Analysis Reveals the Identity of Abeta-Metal Complex Responsible for the Initial Aggregation of Abeta in the Synapse. ACS Chem. Neurosci. 2017, 8, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.F.; Ming, L.J. Metallo-ROS in Alzheimer’s disease: Oxidation of neurotransmitters by CuII-beta-amyloid and neuropathology of the disease. Angew. Chem. Int. Ed. 2007, 46, 3337–3341. [Google Scholar] [CrossRef]
- Nam, E.; Derrick, J.S.; Lee, S.; Kang, J.; Han, J.; Lee, S.J.C.; Chung, S.W.; Lim, M.H. Regulatory Activities of Dopamine and Its Derivatives toward Metal-Free and Metal-Induced Amyloid-beta Aggregation, Oxidative Stress, and Inflammation in Alzheimer’s Disease. ACS Chem. Neurosci. 2018, 9, 2655–2666. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Wezynfeld, N.E.; Stefaniak, E.; Pomorski, A.; Plonka, D.; Krezel, A.; Bal, W.; Faller, P. Cu transfer from amyloid-beta4-16 to metallothionein-3: The role of the neurotransmitter glutamate and metallothionein-3 Zn(ii)-load states. Chem. Commun. 2018, 54, 12634–12637. [Google Scholar] [CrossRef]
- Margerum, D.W.; Dukes, G.R. Metal Ions in Biological Systems. In Metal Ions in Biological Systems; Sigel, H., Ed.; Marcel Dekker: New York, NY, USA, 1974; Volume 1, pp. 157–212. [Google Scholar]
- Ma, Q.F.; Li, Y.M.; Du, J.T.; Kanazawa, K.; Nemoto, T.; Nakanishi, H.; Zhao, Y.F. Binding of copper (II) ion to an Alzheimer’s tau peptide as revealed by MALDI-TOF MS, CD, and NMR. Biopolymers 2005, 79, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, M.; Szunyog, G.; Grenacs, A.; Lihi, N.; Kallay, C.; Di Natale, G.; Campagna, T.; Lanza, V.; Tabbi, G.; Pappalardo, G.; et al. Copper(II) Coordination Abilities of the Tau Protein’s N-Terminus Peptide Fragments: A Combined Potentiometric, Spectroscopic and Mass Spectrometric Study. ChemPlusChem 2019, 84, 1697–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacchella, C.; Gentili, S.; Bellotti, D.; Quartieri, E.; Draghi, S.; Baratto, M.C.; Remelli, M.; Valensin, D.; Monzani, E.; Nicolis, S.; et al. Binding and Reactivity of Copper to R1 and R3 Fragments of tau Protein. Inorg. Chem. 2020, 59, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Di Vaira, M.; Bazzicalupi, C.; Orioli, P.; Messori, L.; Bruni, B.; Zatta, P. Clioquinol, a drug for Alzheimer’s disease specifically interfering with brain metal metabolism: Structural characterization of its zinc(II) and copper(II) complexes. Inorg. Chem. 2004, 43, 3795–3797. [Google Scholar] [CrossRef]
- Chen, D.; Cui, Q.C.; Yang, H.; Barrea, R.A.; Sarkar, F.H.; Sheng, S.; Yan, B.; Reddy, G.P.; Dou, Q.P. Clioquinol, a therapeutic agent for Alzheimer’s disease, has proteasome-inhibitory, androgen receptor-suppressing, apoptosis-inducing, and antitumor activities in human prostate cancer cells and xenografts. Cancer Res. 2007, 67, 1636–1644. [Google Scholar] [CrossRef] [Green Version]
- Tahmasebinia, F.; Emadi, S. Effect of metal chelators on the aggregation of b-amyloid peptides in the presence of copper and iron. Biometals 2017, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Pushie, M.J.; Nienaber, K.H.; Summers, K.L.; Cotelesage, J.J.; Ponomarenko, O.; Nichol, H.K.; Pickering, I.J.; George, G.N. The solution structure of the copper clioquinol complex. J. Inorg. Biochem. 2014, 133, 50–56. [Google Scholar] [CrossRef]
- Opazo, C.; Luza, S.; Villemagne, V.L.; Volitakis, I.; Rowe, C.; Barnham, K.J.; Strozyk, D.; Masters, C.L.; Cherny, R.A.; Bush, A.I. Radioiodinated clioquinol as a biomarker for b-amyloid: Zn complexes in Alzheimer’s disease. Aging Cell 2006, 5, 69–79. [Google Scholar] [CrossRef]
- Pretsch, D.; Rollinger, J.M.; Schmid, A.; Genov, M.; Wohrer, T.; Krenn, L.; Moloney, M.; Kasture, A.; Hummel, T.; Pretsch, A. Prolongation of metallothionein induction combats ass and a-synuclein toxicity in aged transgenic Caenorhabditis elegans. Sci. Rep. 2020, 10, 11707. [Google Scholar] [CrossRef]
- Barnham, K.J.; Ciccotosto, G.D.; Tickler, A.K.; Ali, F.E.; Smith, D.G.; Williamson, N.A.; Lam, Y.H.; Carrington, D.; Tew, D.; Kocak, G.; et al. Neurotoxic, redox-competent Alzheimer’s b-amyloid is released from lipid membrane by methionine oxidation. J. Biol. Chem. 2003, 278, 42959–42965. [Google Scholar] [CrossRef] [Green Version]
- Cherny, R.A.; Ayton, S.; Finkelstein, D.I.; Bush, A.I.; McColl, G.; Massa, S.M. PBT2 Reduces Toxicity in a C. elegans model of polyQ aggregation and extends lifespan, reduces striatal atrophy and improves motor Performance in the R6/2 mouse model of Huntington’s disease. J. Huntingt. Dis. 2012, 1, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Adlard, P.A.; Cherny, R.A.; Finkelstein, D.I.; Gautier, E.; Robb, E.; Cortes, M.; Volitakis, I.; Liu, X.; Smith, J.P.; Perez, K.; et al. Rapid restoration of cognition in Alzheimer’s transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Ab. Neuron 2008, 59, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Bush, A.I.; Tanzi, R.E. Therapeutics for Alzheimer’s disease based on the metal hypothesis. Neurotherapeutics 2008, 5, 421–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, M.; Vendier, L.; Stigliani, J.L.; Meunier, B.; Robert, A. Structures of the copper and zinc complexes of PBT2, a chelating agent evaluated as potential drug for neurodegenerative diseases. Eur. J. Inorg. Chem. 2017, 2017, 600–608. [Google Scholar] [CrossRef]
- Lannfelt, L.; Blennow, K.; Zetterberg, H.; Batsman, S.; Ames, D.; Harrison, J.; Masters, C.L.; Targum, S.; Bush, A.I.; Murdoch, R.; et al. Safety, efficacy, and biomarker findings of PBT2 in targeting Ab as a modifying therapy for Alzheimer’s disease: A phase IIa, double-blind, randomised, placebo-controlled trial. Lancet. Neurol. 2008, 7, 779–786. [Google Scholar] [CrossRef]
- Faux, N.G.; Ritchie, C.W.; Gunn, A.; Rembach, A.; Tsatsanis, A.; Bedo, J.; Harrison, J.; Lannfelt, L.; Blennow, K.; Zetterberg, H.; et al. PBT2 rapidly improves cognition in Alzheimer’s Disease: Additional phase II analyses. J. Alzheimers Dis. 2010, 20, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villemagne, V.L.; Rowe, C.C.; Barnham, K.J.; Cherny, R.; Woodward, M.; Bozinosvski, S.; Salvado, O.; Bourgeat, P.; Perez, K.; Fowler, C.; et al. A randomized, exploratory molecular imaging study targeting amyloid beta with a novel 8-OH quinoline in Alzheimer’s disease: The PBT2-204 IMAGINE study. Alzheimers Dement. 2017, 3, 622–635. [Google Scholar] [CrossRef]
- Kozak, A.; Shapiro, I. Lipophilic Diesters of Chelating Agents. U.S. Patent WO 99/16741, 1 July 1999. [Google Scholar]
- Lee, J.Y.; Friedman, J.E.; Angel, I.; Kozak, A.; Koh, J.Y. The lipophilic metal chelator DP-109 reduces amyloid pathology in brains of human beta-amyloid precursor protein transgenic mice. Neurobiol. Aging 2004, 25, 1315–1321. [Google Scholar] [CrossRef]
- Petri, S.; Calingasan, N.Y.; Alsaied, O.A.; Wille, E.; Kiaei, M.; Friedman, J.E.; Baranova, O.; Chavez, J.C.; Beal, M.F. The lipophilic metal chelators DP-109 and DP-460 are neuroprotective in a transgenic mouse model of amyotrophic lateral sclerosis. J. Neurochem. 2007, 102, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Raz, L.; Yang, Y.; Thompson, J.; Hobson, S.; Pesko, J.; Mobashery, S.; Chang, M.; Rosenberg, G. MMP-9 inhibitors impair learning in spontaneously hypertensive rats. PLoS ONE 2018, 13, e0208357. [Google Scholar] [CrossRef]
- Chen, D.; Darabedian, N.; Li, Z.; Kai, T.; Jiang, D.; Zhou, F. An improved Bathocuproine assay for accurate valence identification and quantification of copper bound by biomolecules. Anal. Biochem. 2016, 497, 27–35. [Google Scholar] [CrossRef]
- Haas, K.L.; Franz, K.J. Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 2009, 109, 4921–4960. [Google Scholar] [CrossRef] [Green Version]
- La Mendola, D. Nerve growth factor catches copper in neuronal inning. Neural Regen. Res. 2020, 15, 665–666. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, J.K. Fluoxetine Induces Apoptotic and Oxidative Neuronal Death Associated with The Influx of Copper Ions in Cultured Neuronal Cells. Chonnam Med. J. 2020, 56, 20–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walshe, J.M. Penicillamine, a new oral therapy for Wilson’s disease. Am. J. Med. 1956, 21, 487–495. [Google Scholar] [CrossRef]
- Walshe, J.M. Copper chelation in patients with Wilson’s disease. A comparison of penicillamine and triethylene tetramine dihydrochloride. Q. J. Med. 1973, 42, 441–452. [Google Scholar] [PubMed]
- Brewer, G.J. Zinc and tetrathiomolybdate for the treatment of Wilson’s disease and the potential efficacy of anticopper therapy in a wide variety of diseases. Metallomics 2009, 1, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, C.; Bronstein, J.M. Wilson Disease: An Overview and Approach to Management. Neurol. Clin. 2020, 38, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Kou, H.; Zhao, P.; Zheng, W.; Xu, H.; Zhang, X.; Lan, W.; Guo, C.; Wang, T.; Guo, F.; et al. Nasal delivery of D-penicillamine hydrogel upregulates a disintegrin and metalloprotease 10 expression via melatonin receptor 1 in Alzheimer’s disease models. Front. Aging Neurosci. 2021, 13, 660249. [Google Scholar] [CrossRef]
- Cooper, G.J. Therapeutic potential of copper chelation with triethylenetetramine in managing diabetes mellitus and Alzheimer’s disease. Drugs 2011, 71, 128–1320. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Castoldi, F.; Zischka, H.; Kroemer, G. Extending the mode of action of triethylenetetramine (trientine): Autophagy besides copper chelation. J. Hepatol. 2020, 73, 970–972. [Google Scholar] [CrossRef]
- Brewer, G.J.; Dick, R.D.; Yuzbasiyan-Gurkin, V.; Tankanow, R.; Young, A.B.; Kluin, K.J. Initial therapy of patients with Wilson’s disease with tetrathiomolybdate. Arch. Neurol. 1991, 48, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.H.; Guo, C.; Gao, H.L.; Zhong, M.L.; Huang, T.T.; Liu, N.N.; Guo, R.F.; Lan, T.; Zhang, W.; et al. Tetrathiomolybdate Treatment Leads to the Suppression of Inflammatory Responses through the TRAF6/NFkappaB Pathway in LPS-Stimulated BV-2 Microglia. Front. Againg Neurosci. 2018, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Brewer, G.J.; Askari, F.; Lorincz, M.T.; Carlson, M.; Schilsky, M.; Kluin, K.J.; Hedera, P.; Moretti, P.; Fink, J.K.; Tankanow, R.; et al. Treatment of Wilson disease with ammonium tetrathiomolybdate: IV. Comparison of tetrathiomolybdate and trientine in a double-blind study of treatment of the neurologic presentation of Wilson disease. Arch. Neurol. 2006, 63, 521–527. [Google Scholar] [CrossRef]
- Dickens, M.G.; Franz, K.J. A prochelator activated by hydrogen peroxide prevents metal-induced amyloid Beta aggregation. Chembiochem Eur. J. Chem. Biol. 2010, 11, 59–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avramovich-Tirosh, Y.; Amit, T.; Bar-Am, O.; Zheng, H.; Fridkin, M.; Youdim, M.B. Therapeutic targets and potential of the novel brain- permeable multifunctional iron chelator-monoamine oxidase inhibitor drug, M-30, for the treatment of Alzheimer’s disease. J. Neurochem. 2007, 100, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Mechlovich, D.; Amit, T.; Mandel, S.A.; Bar-Am, O.; Bloch, K.; Vardi, P.; Youdim, M.B. The novel multifunctional, iron-chelating drugs M30 and HLA20 protect pancreatic b-cell lines from oxidative stress damage. J. Pharmacol. Exp. Ther. 2010, 333, 874–882. [Google Scholar] [CrossRef]
- Zheng, H.; Youdim, M.B.; Fridkin, M. Site-activated chelators targeting acetylcholinesterase and monoamine oxidase for Alzheimer’s therapy. ACS Chem. Biol. 2010, 5, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Youdim, M.B.; Fridkin, M. Site-activated multifunctional chelator with acetylcholinesterase and neuroprotective-neurorestorative moieties for Alzheimer’s therapy. J. Med. Chem. 2009, 52, 4095–4098. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.M.; Vieira, R.P.; Jones, M.R.; Wang, M.C.; Dyrager, C.; Souza-Fagundes, E.M.; Da Silva, J.G.; Storr, T.; Beraldo, H. 8-Hydroxyquinoline Schiff-base compounds as antioxidants and modulators of copper-mediated Ab peptide aggregation. J. Inorg. Biochem. 2014, 139, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Hu, J.; Yang, X.; Feng, X.; Li, X.; Huang, L.; Chan, A.S.C. Design, synthesis, and evaluation of orally bioavailable quinoline-indole derivatives as innovative multitarget-directed ligands: Promotion of cell proliferation in the adult murine hippocampus for the treatment of Alzheimer’s disease. J. Med. Chem. 2018, 61, 1871–1894. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, V.; Puglisi, A.; Viale, M.; Aiello, C.; Sgarlata, C.; Vecchio, G.; Clarke, J.; Milton, J.; Spencer, J. New cyclodextrin-bearing 8-hydroxyquinoline ligands as multifunctional molecules. Chem. Eur. J. 2013, 19, 13946–13955. [Google Scholar] [CrossRef] [PubMed]
- Groenning, M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J. Chem. Biol. 2010, 3, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dedeoglu, A.; Cormier, K.; Payton, S.; Tseitlin, K.A.; Kremsky, J.N.; Lai, L.; Li, X.; Moir, R.D.; Tanzi, R.E.; Bush, A.I.; et al. Preliminary studies of a novel bifunctional metal chelator targeting Alzheimer’s amyloidogenesis. Exp. Gerontol. 2004, 39, 1641–1649. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, C.; Sanchez de Groot, N.; Rimola, A.; Alvarez-Larena, A.; Lloveras, V.; Vidal-Gancedo, J.; Ventura, S.; Vendrell, J.; Sodupe, M.; Gonzalez-Duarte, P. Design, selection, and characterization of thioflavin-based intercalation compounds with metal chelating properties for application in Alzheimer’s disease. J. Am. Chem. Soc. 2009, 131, 1436–1451. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, C.; Telpoukhovskaia, M.A.; Ali-Torres, J.; Rodriguez-Santiago, L.; Manso, Y.; Bailey, G.A.; Hidalgo, J.; Sodupe, M.; Orvig, C. Thioflavin-based molecular probes for application in Alzheimer’s disease: From in silico to in vitro models. Metallomics 2015, 7, 83–92. [Google Scholar] [CrossRef]
- Barabash, R.D.; Skobelkin, O.K.; Petukhov, M.I.; Normanskii, V.E.; Riazhskii, G.G.; Mironova, M.I.; Andreeva, K.P.; Kolobanov, A.S. The pharmacokinetics of hematoporphyrin, its derivative and fluorescein during the development of carcinosarcoma. Farmakol. I Toksikol. 1990, 53, 24–26. [Google Scholar]
- Sharma, A.K.; Pavlova, S.T.; Kim, J.; Finkelstein, D.; Hawco, N.J.; Rath, N.P.; Kim, J.; Mirica, L.M. Bifunctional compounds for controlling metal-mediated aggregation of the Ab42 peptide. J. Am.Chem.Soc. 2012, 134, 6625–6636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, J.; Li, M.; Wu, L.; Ren, J.; Qu, X. Liberation of copper from amyloid plaques: Making a risk factor useful for Alzheimer’s disease treatment. J. Med. Chem. 2012, 55, 9146–9155. [Google Scholar] [CrossRef]
- Jones, M.R.; Mu, C.; Wang, M.C.; Webb, M.I.; Walsby, C.J.; Storr, T. Modulation of the Ab peptide aggregation pathway by KP1019 limits Ab-associated neurotoxicity. Metallomics 2015, 7, 129–135. [Google Scholar] [CrossRef]
- Hindo, S.S.; Mancino, A.M.; Braymer, J.J.; Liu, Y.; Vivekanandan, S.; Ramamoorthy, A.; Lim, M.H. Small molecule modulators of copper-induced Ab aggregation. J. Am. Chem. Soc. 2009, 131, 16663–16665. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Braymer, J.J.; Nanga, R.P.; Ramamoorthy, A.; Lim, M.H. Design of small molecules that target metal-Ab species and regulate metal-induced Ab aggregation and neurotoxicity. Proc. Natl. Acad. Sci. USA 2010, 107, 21990–21995. [Google Scholar] [CrossRef] [Green Version]
- Geldenhuys, W.J.; Ko, K.S.; Stinnett, H.; Schyf, C.J.V.d.; Lim, M.H. Identification of multifunctional small molecule-based reversible monoamine oxidase inhibitors. MedChemComm 2011, 2, 1099–1103. [Google Scholar] [CrossRef]
- Savelieff, M.G.; Liu, Y.; Senthamarai, R.R.; Korshavn, K.J.; Lee, H.J.; Ramamoorthy, A.; Lim, M.H. A small molecule that displays marked reactivity toward copper- versus zinc-amyloid-b implicated in Alzheimer’s disease. Chem. Commun. 2014, 50, 5301–5303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Zheng, X.; Krishnamoorthy, J.; Savelieff, M.G.; Park, H.M.; Brender, J.R.; Kim, J.H.; Derrick, J.S.; Kochi, A.; Lee, H.J.; et al. Rational design of a structural framework with potential use to develop chemical reagents that target and modulate multiple facets of Alzheimer’s disease. J. Am. Chem. Soc. 2014, 136, 299–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, M.W.; Derrick, J.S.; Kerr, R.A.; Oh, S.B.; Cho, W.J.; Lee, S.J.; Ji, Y.; Han, J.; Tehrani, Z.A.; Suh, N.; et al. Structure-mechanism-based engineering of chemical regulators targeting distinct pathological factors in Alzheimer’s disease. Nat. Commun. 2016, 7, 13115. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.W.; Derrick, J.S.; Suh, J.M.; Kim, M.; Korshavn, K.J.; Kerr, R.A.; Cho, W.J.; Larsen, S.D.; Ruotolo, B.T.; Ramamoorthy, A.; et al. Minor Structural Variations of Small Molecules Tune Regulatory Activities toward Pathological Factors in Alzheimer’s Disease. ChemMedChem 2017, 12, 1828–1838. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Korshavn, K.J.; Nam, Y.; Kang, J.; Paul, T.J.; Kerr, R.A.; Youn, I.S.; Ozbil, M.; Kim, K.S.; Ruotolo, B.T.; et al. Structural and mechanistic insights into development of chemical tools to control individual and inter-related pathological features in Alzheimer’s disease. Chem. Eur. J. 2017, 23, 2706–2715. [Google Scholar] [CrossRef] [Green Version]
- Guan, L.; Hao, Y.; Chen, L.; Wei, M.-L.; Jiang, Q.; Liu, W.-Y.; Zhang, Y.-B.; Zhang, J.; Feng, F.; Qu, W. Synthesis and evaluation of neuroprotective 4-O-substituted chrysotoxine derivatives as potential multifunctional agents for the treatment of Alzheimer’s disease. RSC Adv. 2016, 6, 22827–22838. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, X.-B.; Li, Z.-R.; Li, S.-Y.; Xie, S.-S.; Huanga, M.; Kong, L.-Y. Design of a structural framework with potential use to develop balanced multifunctional agents against Alzheimer’s disease. RSC Adv. 2015, 5, 14242–14255. [Google Scholar] [CrossRef]
- Jones, M.R.; Dyrager, C.; Hoarau, M.; Korshavn, K.J.; Lim, M.H.; Ramamoorthy, A.; Storr, T. Multifunctional quinoline-triazole derivatives as potential modulators of amyloid-b peptide aggregation. J. Inorg. Biochem. 2016, 158, 131–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.R.; Mathieu, E.; Dyrager, C.; Faissner, S.; Vaillancourt, Z.; Korshavn, K.J.; Lim, M.H.; Ramamoorthy, A.; Wee Yong, V.; Tsutsui, S.; et al. Multi-target-directed phenol-triazole ligands as therapeutic agents for Alzheimer’s disease. Chem. Sci. 2017, 8, 5636–5643. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Gomes, L.M.F.; Zhang, T.; Storr, T. A small bifunctional chelator that modulates Ab42 aggregation. Can. J. Chem. 2017, 96, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Chen, J.; Li, X.; Su, T.; Wang, Y.; Wang, Z.; Huang, L.; Li, X. Synthesis and evaluation of selegiline derivatives as monoamine oxidase inhibitor, antioxidant and metal chelator against Alzheimer’s disease. Bioorg. Med. Chem. 2015, 23, 3722–3729. [Google Scholar] [CrossRef]
- Li, Y.; Qiang, X.; Luo, L.; Li, Y.; Xiao, G.; Tan, Z.; Deng, Y. Synthesis and evaluation of 4-hydroxyl aurone derivatives as multifunctional agents for the treatment of Alzheimer’s disease. Bioorg. Med. Chem. 2016, 24, 2342–2351. [Google Scholar] [CrossRef]
- Li, F.; Wu, J.J.; Wang, J.; Yang, X.L.; Cai, P.; Liu, Q.H.; Kong, L.Y.; Wang, X.B. Synthesis and pharmacological evaluation of novel chromone derivatives as balanced multifunctional agents against Alzheimer’s disease. Bioorg. Med. Chem. 2017, 25, 3815–3826. [Google Scholar] [CrossRef]
- Li, S.Y.; Wang, X.B.; Kong, L.Y. Design, synthesis and biological evaluation of imine resveratrol derivatives as multi-targeted agents against Alzheimer’s disease. Eur. J. Med. Chem. 2014, 71, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, M.; Sheng, R.; Ma, Y. Synthesis and biological evaluation of deferiprone-resveratrol hybrids as antioxidants, Ab1-42 aggregation inhibitors and metal-chelating agents for Alzheimer’s disease. Eur. J. Med. Chem. 2017, 127, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.R.; Ayton, S.; Bush, A.I. Iron and Alzheimer’s Disease: An Update on Emerging Mechanisms. J. Alzheimers Dis. 2018, 64, S379–S395. [Google Scholar] [CrossRef] [PubMed]
- Mason, H.S. Mechanisms of oxygen metabolism. Science 1957, 125, 1185–1188. [Google Scholar] [CrossRef] [PubMed]
- Lieu, P.T.; Heiskala, M.; Peterson, P.A.; Yang, Y. The roles of iron in health and disease. Mol. Asp. Med. 2001, 22, 1–87. [Google Scholar] [CrossRef]
- Nunez, M.T.; Chana-Cuevas, P. New perspectives in iron chelation therapy for the treatment of neurodegenerative diseases. Pharmaceuticals 2018, 11, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiban, J.; So, M.; Kaguni, L.S. Iron-sulfur clusters in mitochondrial metabolism: Multifaceted roles of a simple cofactor. Biochemistry 2016, 81, 1066–1080. [Google Scholar] [CrossRef]
- Que, E.L.; Domaille, D.W.; Chang, C.J. Metals in neurobiology: Probing their chemistry and biology with molecular imaging. Chem. Rev. 2008, 108, 1517–1549. [Google Scholar] [CrossRef]
- Belaidi, A.A.; Bush, A.I. Iron neurochemistry in Alzheimer’s disease and Parkinson’s disease: Targets for therapeutics. J. Neurochem. 2016, 139, 179–197. [Google Scholar] [CrossRef] [Green Version]
- Nnah, I.C.; Wessling-Resnick, M. Brain Iron Homeostasis: A Focus on Microglial Iron. Pharmaceuticals 2018, 11, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, D.J.; Merlot, A.M.; Huang, M.L.; Bae, D.H.; Jansson, P.J.; Sahni, S.; Kalinowski, D.S.; Richardson, D.R. Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta 2015, 1853, 1130–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haywood, S.; Paris, J.; Ryvar, R.; Botteron, C. Brain copper elevation and neurological changes in north ronaldsay sheep: A model for neurodegenerative disease? J. Comp. Pathol. 2008, 139, 252–255. [Google Scholar] [CrossRef]
- Gerlach, M.; Ben-Shachar, D.; Riederer, P.; Youdim, M.B. Altered brain metabolism of iron as a cause of neurodegenerative diseases? J. Neurochem. 1994, 63, 793–807. [Google Scholar] [CrossRef]
- Hentze, M.W.; Muckenthaler, M.U.; Andrews, N.C. Balancing acts: Molecular control of mammalian iron metabolism. Cell 2004, 117, 285–297. [Google Scholar] [CrossRef] [Green Version]
- Whitnall, M.; Suryo Rahmanto, Y.; Huang, M.L.; Saletta, F.; Lok, H.C.; Gutierrez, L.; Lazaro, F.J.; Fleming, A.J.; St Pierre, T.G.; Mikhael, M.R.; et al. Identification of nonferritin mitochondrial iron deposits in a mouse model of Friedreich ataxia. Proc. Natl. Acad. Sci. USA 2012, 109, 20590–20595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, G.M.; Dang, T.N.; Dringen, R.; Robinson, S.R. Accumulation of non-transferrin-bound iron by neurons, astrocytes, and microglia. Neurotox. Res. 2011, 19, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Dringen, R.; Bishop, G.M.; Koeppe, M.; Dang, T.N.; Robinson, S.R. The pivotal role of astrocytes in the metabolism of iron in the brain. Neurochem. Res. 2007, 32, 1884–1890. [Google Scholar] [CrossRef] [PubMed]
- Moos, T.; Rosengren Nielsen, T.; Skjorringe, T.; Morgan, E.H. Iron trafficking inside the brain. J. Neurochem. 2007, 103, 1730–1740. [Google Scholar] [CrossRef]
- Ponka, P.; Beaumont, C.; Richardson, D.R. Function and regulation of transferrin and ferritin. Semin. Hematol. 1998, 35, 35–54. [Google Scholar]
- Salvador, G.A. Iron in neuronal function and dysfunction. BioFactors 2010, 36, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, C.; Di Pisa, F.; Bernacchioni, C.; Ciambellotti, S.; Turano, P.; Mangani, S. Iron binding to human heavy-chain ferritin. Acta Crystallogr. D 2015, 71, 1909–1920. [Google Scholar] [CrossRef]
- Pfaffen, S.; Abdulqadir, R.; Le Brun, N.E.; Murphy, M.E. Mechanism of ferrous iron binding and oxidation by ferritin from a pennate diatom. J. Biol. Chem. 2013, 288, 14917–14925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffen, S.; Bradley, J.M.; Abdulqadir, R.; Firme, M.R.; Moore, G.R.; Le Brun, N.E.; Murphy, M.E.P. A diatom ferritin optimized for iron oxidation but not iron storage. J. Biol. Chem. 2015, 290, 28416–28427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozzi, C.; Ciambellotti, S.; Bernacchioni, C.; Di Pisa, F.; Mangani, S.; Turano, P. Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional (m3-oxo)Tris[(m2-peroxo)] triiron(III) cluster. Proc. Natl. Acad. Sci. USA 2017, 114, 2580–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, T.E.; Mason, A.B.; He, Q.Y.; Halbrooks, P.J.; Briggs, S.K.; Smith, V.C.; MacGillivray, R.T.; Everse, S.J. The position of arginine 124 controls the rate of iron release from the N-lobe of human serum transferrin. A structural study. J. Biol. Chem. 2003, 278, 6027–6033. [Google Scholar]
- Mason, A.B.; Halbrooks, P.J.; James, N.G.; Connolly, S.A.; Larouche, J.R.; Smith, V.C.; MacGillivray, R.T.; Chasteen, N.D. Mutational analysis of C-lobe ligands of human serum transferrin: Insights into the mechanism of iron release. Biochemistry 2005, 44, 8013–8021. [Google Scholar] [CrossRef]
- McCarthy, R.C.; Kosman, D.J. Iron transport across the blood-brain barrier: Development, neurovascular regulation and cerebral amyloid angiopathy. Cell. Mol. Life Sci. 2015, 72, 709–727. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.; Moreton, K. The distribution of iron between the metal-binding sites of transferrin human serum. Biochem. J. 1980, 185, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Connor, J.R.; Fine, R.E. The distribution of transferrin immunoreactivity in the rat central nervous system. Brain Res. 1986, 368, 319–328. [Google Scholar] [CrossRef]
- Masaldan, S.; Bush, A.I.; Devos, D.; Rolland, A.S.; Moreau, C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free Radic. Biol. Med. 2019, 133, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, A.; Skjorringe, T.; Johnsen, K.B.; Siupka, P.; Thomsen, L.B.; Nielsen, M.S.; Thomsen, L.L.; Moos, T. Expression of iron-related proteins at the neurovascular unit supports reduction and reoxidation of iron for transport through the blood-brain barrier. Mol. Neurobiol. 2016, 53, 7237–7253. [Google Scholar] [CrossRef]
- Pinero, D.J.; Connor, J.R. Iron in the brain: An important contributor in normal and diseased states. Neuroscientist 2000, 6, 435–453. [Google Scholar] [CrossRef]
- Skjorringe, T.; Burkhart, A.; Johnsen, K.B.; Moos, T. Divalent metal transporter 1 (DMT1) in the brain: Implications for a role in iron transport at the blood-brain barrier, and neuronal and glial pathology. Front. Mol. Neurosci. 2015, 8, 19. [Google Scholar] [PubMed] [Green Version]
- Garrick, M.D.; Singleton, S.T.; Vargas, F.; Kuo, H.C.; Zhao, L.; Knopfel, M.; Davidson, T.; Costa, M.; Paradkar, P.; Roth, J.A.; et al. DMT1: Which metals does it transport? Biol. Res. 2006, 39, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liuzzi, J.P.; Aydemir, F.; Nam, H.; Knutson, M.D.; Cousins, R.J. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natal. Acad. Sci. USA 2006, 103, 13612–13617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkitkasemwong, S.; Wang, C.Y.; Mackenzie, B.; Knutson, M.D. Physiologic implications of metal-ion transport by ZIP14 and ZIP8. Biometals 2012, 25, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Andrews, N.C. The iron transporter DMT1. Int. J. Biochem. Cell Biol. 1999, 31, 991–994. [Google Scholar] [CrossRef]
- Wang, C.Y.; Jenkitkasemwong, S.; Duarte, S.; Sparkman, B.K.; Shawki, A.; Mackenzie, B.; Knutson, M.D. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J. Biol. Chem. 2012, 287, 34032–34043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, N.; Gao, J.; Enns, C.A.; Knutson, M.D. ZRT/IRT-like protein 14 (ZIP14) promotes the cellular assimilation of iron from transferrin. J. Biol. Chem. 2010, 285, 32141–32150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, A.E.; Mendez, M.J.; Hokanson, C.A.; Rees, D.C.; Bjorkman, P.J. Investigation of the biophysical and cell biological properties of ferroportin, a multipass integral membrane protein iron exporter. J. Mol. Biol. 2009, 386, 717–732. [Google Scholar] [CrossRef] [Green Version]
- Ward, D.M.; Kaplan, J. Ferroportin-mediated iron transport: Expression and regulation. Biochim. Biophys. Acta 2012, 1823, 1426–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.Y.; David, S. Glycosylphosphatidylinositol-anchored ceruloplasmin is required for iron efflux from cells in the central nervous system. J. Biol. Chem. 2003, 278, 27144–27148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Z.M.; Shen, X. Brain iron transport and neurodegeneration. Trends Mol. Med. 2001, 7, 103–108. [Google Scholar] [CrossRef]
- Qian, Z.M.; Wang, Q. Expression of iron transport proteins and excessive iron accumulation in the brain in neurodegenerative disorders. Brain Res. Rev. 1998, 27, 257–267. [Google Scholar] [PubMed]
- Malecki, E.A.; Devenyi, A.G.; Beard, J.L.; Connor, J.R. Existing and emerging mechanisms for transport of iron and manganese to the brain. J. Neurosci. Res. 1999, 56, 113–122. [Google Scholar] [CrossRef]
- Nam, G.; Yi, Y.; Lee, H.J.; Lee, J.; Kang, J.; Lim, M.H. Metalloneurochemistry. In Comprehensive Coordination Chemistry, 3rd ed.; Constable, E., Parkin, G., Que, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar] [CrossRef]
- Beard, J.L. Iron biology in immune function, muscle metabolism and neuronal functioning. J. Nutr. 2001, 131, 568S–579S. [Google Scholar] [CrossRef] [PubMed]
- Singh, N. The role of iron in prion disease and other neurodegenerative diseases. PLoS Pathog. 2014, 10, e1004335. [Google Scholar] [CrossRef]
- Agarwal, K.N. Iron and the brain: Neurotransmitter receptors and magnetic resonance spectroscopy. Br. J. Nutr. 2001, 85, S147–S150. [Google Scholar] [CrossRef]
- Magistretti, P.J.; Allaman, I. A cellular perspective on brain energy metabolism and functional imaging. Neuron 2015, 86, 883–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lill, R.; Hoffmann, B.; Molik, S.; Pierik, A.J.; Rietzschel, N.; Stehling, O.; Uzarska, M.A.; Webert, H.; Wilbrecht, C.; Muhlenhoff, U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 2012, 1823, 1491–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, E.; Pasquini, J.M.; Thompson, K.; Felt, B.; Butkus, G.; Beard, J.; Connor, J.R. Effect of manipulation of iron storage, transport, or availability on myelin composition and brain iron content in three different animal models. J. Neurosci. Res. 2004, 77, 681–689. [Google Scholar] [CrossRef]
- Yu, G.S.; Steinkirchner, T.M.; Rao, G.A.; Larkin, E.C. Effect of prenatal iron deficiency on myelination in rat pups. Am. J. Pathol. 1986, 125, 620–624. [Google Scholar] [PubMed]
- Connor, J.R.; Menzies, S.L. Relationship of iron to oligodendrocytes and myelination. Glia 1996, 17, 83–93. [Google Scholar] [CrossRef]
- Kim, J.; Wessling-Resnick, M. Iron and mechanisms of emotional behavior. J. Nutr. Biochem. 2014, 25, 1101–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwik-Uribe, C.L.; Gietzen, D.; German, J.B.; Golub, M.S.; Keen, C.L. Chronic marginal iron intakes during early development in mice result in persistent changes in dopamine metabolism and myelin composition. J. Nutr. 2000, 130, 2821–2830. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, D.M.; Ruskin, B.; Lovenberg, W. Tryptophan hydroxylase. The role of oxygen, iron, and sulfhydryl groups as determinants of stability and catalytic activity. J. Biol. Chem. 1980, 255, 4137–4143. [Google Scholar] [CrossRef]
- Bradbury, M.W. Transport of iron in the blood-brain-cerebrospinal fluid system. J. Neurochem. 1997, 69, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Sayre, L.M.; Perry, G.; Harris, P.L.; Liu, Y.; Schubert, K.A.; Smith, M.A. In situ oxidative catalysis by neurofibrillary tangles and senile plaques in Alzheimer’s disease: A central role for bound transition metals. J. Neurochem. 2000, 74, 270–279. [Google Scholar] [CrossRef]
- Suh, S.W.; Jensen, K.B.; Jensen, M.S.; Silva, D.S.; Kesslak, P.J.; Danscher, G.; Frederickson, C.J. Histochemically-reactive zinc in amyloid plaques, angiopathy, and degenerating neurons of Alzheimer’s diseased brains. Brain Res. 2000, 852, 274–278. [Google Scholar] [CrossRef]
- Bartzokis, G.; Sultzer, D.; Cummings, J.; Holt, L.E.; Hance, D.B.; Henderson, V.W.; Mintz, J. In vivo evaluation of brain iron in Alzheimer disease using magnetic resonance imaging. Arch. Gen. Psychiatry 2000, 57, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovell, M.A.; Robertson, J.D.; Teesdale, W.J.; Campbell, J.L.; Markesbery, W.R. Copper, iron and zinc in Alzheimer’s disease senile plaques. J. Neurol. Sci. 1998, 158, 47–52. [Google Scholar] [CrossRef]
- Yamamoto, A.; Shin, R.W.; Hasegawa, K.; Naiki, H.; Sato, H.; Yoshimasu, F.; Kitamoto, T. Iron (III) induces aggregation of hyperphosphorylated tau and its reduction to iron (II) reverses the aggregation: Implications in the formation of neurofibrillary tangles of Alzheimer’s disease. J. Neurochem. 2002, 82, 1137–1147. [Google Scholar] [CrossRef]
- Huang, X.; Moir, R.D.; Tanzi, R.E.; Bush, A.I.; Rogers, J.T. Redox active metals, oxidative stress, and Alzheimer’s disease pathology. Ann. N. Y. Acad. Sci. 2004, 1012, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Chevion, M. The role of iron in beta amyloid toxicity. Biochem. Biophys. Res. Commun. 1995, 216, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Carocci, A.; Catalano, A.; Sinicropi, M.S.; Genchi, G. Oxidative stress and neurodegeneration: The involvement of iron. Biometals 2018, 31, 715–735. [Google Scholar] [CrossRef]
- Uranga, R.M.; Salvador, G.A. Unraveling the burden of iron in neurodegeneration: Intersections with amyloid b peptide pathology. Oxid. Med. Cell Longev. 2018, 2018, 2850341. [Google Scholar] [CrossRef]
- Ward, R.J.; Zucca, F.A.; Duyn, J.H.; Crichton, R.R.; Zecca, L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014, 13, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Bansal, S.; Biswas, G.; Avadhani, N.G. Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol. 2014, 2, 273–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Y.; Luo, M.; Yao, F.; Wang, S.; Yuan, Z.; Yang, Y. Ceruloplasmin suppresses ferroptosis by regulating iron homeostasis in hepatocellular carcinoma cells. Cell. Signal. 2020, 72, 109633. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keberle, H. The Biochemistry of Desferrioxamine and Its Relation to Iron Metabolism. Ann. N.Y. Acad. Sci. 1964, 119, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ma, Y.; Kong, X.; Hider, R.C. Design of iron chelators with therapeutic application. Dalton Trans. 2012, 41, 6371–6389. [Google Scholar] [CrossRef] [PubMed]
- Fredenburg, A.M.; Sethi, R.K.; Allen, D.D.; Yokel, R.A. The pharmacokinetics and blood-brain barrier permeation of the chelators 1,2 dimethly-, 1,2 diethyl-, and 1-[ethan-1’ol]-2-methyl-3-hydroxypyridin-4-one in the rat. Toxicology 1996, 108, 191–199. [Google Scholar] [CrossRef]
- Ben-Shachar, D.; Eshel, G.; Finberg, J.P.; Youdim, M.B. The iron chelator desferrioxamine (Desferal) retards 6-hydroxydopamine-induced degeneration of nigrostriatal dopamine neurons. J. Neurochem. 1991, 56, 1441–1444. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, D.S.; Richardson, D.R. The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol. Rev. 2005, 57, 547–583. [Google Scholar] [CrossRef]
- Evers, A.; Hancock, R.D.; Martell, A.E.; Motekaitis, R.J. Metal ion recognition in ligands with negatively charged oxygen donor groups. Complexation of iron(III), gallium(III), indium(III), aluminum(III), and other highly charged metal ions. Inorg. Chem. 1989, 28, 2189–2195. [Google Scholar] [CrossRef]
- Lan, J.; Jiang, D.H. Desferrioxamine and vitamin E protect against iron and MPTP-induced neurodegeneration in mice. J. Neural Transm. 1997, 104, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Cable, H.; Lloyd, J.B. Cellular uptake and release of two contrasting iron chelators. J. Pharm. Pharmacol. 1999, 51, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, M.L. Deferoxamine enhances alternative activation of microglia and inhibits amyloid beta deposits in APP/PS1 mice. Brain Res. 2017, 1677, 8692. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.M.; Kosyakovsky, J.; Baillargeon, A.M.; Tokarev, J.V.; Cooner, J.M.; Svitak, A.L.; Faltesek, K.A.; Frey, W.H., II; Hanson, L.R. Intranasal deferoxamine can improve memory in healthy C57 mice, suggesting a partially non-disease-specific pathway of functional neurologic improvement. Brain Behav. 2020, 10, e01536. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.M.; Forsberg, A.C.; Stroebel, B.M.; Faltesek, K.A.; Verden, D.R.; Hamel, K.A.; Raney, E.B.; Crow, J.M.; Haase, L.R.; Knutzen, K.E.; et al. Intranasal deferoxamine affects memory loss, oxidation, and the insulin pathway in the streptozotocin rat model of Alzheimer’s disease. J. Neurol. Sci. 2017, 380, 164–171. [Google Scholar] [CrossRef]
- Molina-Holgado, F.; Gaeta, A.; Francis, P.T.; Williams, R.J.; Hider, R.C. Neuroprotective actions of deferiprone in cultured cortical neurones and SHSY-5Y cells. J. Neurochem. 2008, 105, 2466–2476. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.; Marques, S.M.; Quintanova, C.; Silva, D.F.; Cardoso, S.M.; Chaves, S.; Santos, M.A. Multifunctional iron-chelators with protective roles against neurodegenerative diseases. Dalton Trans. 2013, 42, 6058–6073. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Portbury, S.D.; Lago, L.; McColl, G.; Finkelstein, D.I.; Bush, A.I.; Adlard, P.A. The Iron Chelator Deferiprone Improves the Phenotype in a Mouse Model of Tauopathy. J. Alzheimers Dis. 2020, 77, 753–771. [Google Scholar] [CrossRef] [PubMed]
- Prasanthi, J.R.; Schrag, M.; Dasari, B.; Marwarha, G.; Dickson, A.; Kirsch, W.M.; Ghribi, O. Deferiprone reduces amyloid-beta and tau phosphorylation levels but not reactive oxygen species generation in hippocampus of rabbits fed a cholesterol-enriched diet. J. Alzheimers Dis. 2012, 30, 167–182. [Google Scholar] [CrossRef] [Green Version]
- Gleason, A.; Bush, A.I. Iron and ferroptosis as therapeutic targets in Alzheimer’s disease. Neurotherapeutics 2021, 18, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Martin-Bastida, A.; Ward, R.J.; Newbould, R.; Piccini, P.; Sharp, D.; Kabba, C.; Patel, M.C.; Spino, M.; Connelly, J.; Tricta, F.; et al. Brain iron chelation by deferiprone in a phase 2 randomised double-blinded placebo controlled clinical trial in Parkinson’s disease. Sci. Rep. 2017, 7, 1398. [Google Scholar] [CrossRef] [PubMed]
- Mudasir, X.; Arai, M.; Yoshioka, N.; Inoue, H. Reversed-phase high-performance liquid chromatography of iron(II) and copper(II) chelates with 4,7-diphenyl-1,10-phenanthroline disulfonate. J. Chromatogr. A 1998, 799, 171–176. [Google Scholar] [CrossRef]
- Yokoyama, T.; Nakano, K.; Zenki, M. Specific separation of nickel ion with bathophenanthroline disulfonic acid by capillary zone electrophoresis. Anal. Chim. Acta 1999, 396, 117–123. [Google Scholar] [CrossRef]
- Randell, E.W.; Parkes, J.G.; Olivieri, N.F.; Templeton, D.M. Uptake of non-transferrin-bound iron by both reductive and nonreductive processes is modulated by intracellular iron. J. Biol. Chem. 1994, 269, 16046–16053. [Google Scholar] [CrossRef]
- Amit, T.; Bar-Am, O.; Mechlovich, D.; Kupershmidt, L.; Youdim, M.B.H.; Weinreb, O. The novel multitarget iron chelating and propargylamine drug M30 affects APP regulation and processing activities in Alzheimer’s disease models. Neuropharmacology 2017, 123, 359–367. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, P.; Chen, B.; Lun, P.; Dou, Y.; Cao, W.W. A study on the effect of ion chelating agent on Alzheimer disease. Biomed. Res. 2017, 28, 8022–8026. [Google Scholar]
- Peters, C.; Bascunan, D.; Burgos, C.F.; Bobadilla, C.; Gonzalez-Sanmiguel, J.; Boopathi, S.; Riffo, N.; Fernandez-Perez, E.J.; Tarnok, M.E.; Aguilar, L.F.; et al. Characterization of a new molecule capable of inhibiting several steps of the amyloid cascade in Alzheimer’s disease. Neurobiol. Dis. 2020, 141, 104938. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Ho, T.-L. A New Glycosylation Method Based on 8-Quinolyl Glycosides. J. Chin. Chem. Soc. 2006, 53, 1567–1570. [Google Scholar] [CrossRef]
- Zheng, H.; Weiner, L.M.; Bar-Am, O.; Epsztejn, S.; Cabantchik, Z.I.; Warshawsky, A.; Youdim, M.B.; Fridkin, M. Design, synthesis, and evaluation of novel bifunctional iron-chelators as potential agents for neuroprotection in Alzheimer’s, Parkinson’s, and other neurodegenerative diseases. Bioorg. Med. Chem. 2005, 13, 773–783. [Google Scholar] [CrossRef]
- Knez, D.; Brus, B.; Coquelle, N.; Sosic, I.; Sink, R.; Brazzolotto, X.; Mravljak, J.; Colletier, J.P.; Gobec, S. Structure-based development of nitroxoline derivatives as potential multifunctional anti-Alzheimer agents. Bioorg. Med. Chem. 2015, 23, 4442–4452. [Google Scholar] [CrossRef]
- Shachar, D.B.; Kahana, N.; Kampel, V.; Warshawsky, A.; Youdim, M.B. Neuroprotection by a novel brain permeable iron chelator, VK-28, against 6-hydroxydopamine lession in rats. Neuropharmacology 2004, 46, 254–263. [Google Scholar] [CrossRef]
- Cacciatore, I.; Cornacchia, C.; Fornasari, E.; Baldassarre, L.; Pinnen, F.; Sozio, P.; Di Stefano, A.; Marinelli, L.; Dean, A.; Fulle, S.; et al. A glutathione derivative with chelating and in vitro neuroprotective activities: Synthesis, physicochemical properties, and biological evaluation. ChemMedChem 2013, 8, 1818–1829. [Google Scholar] [CrossRef]
- Kupershmidt, L.; Weinreb, O.; Amit, T.; Mandel, S.; Bar-Am, O.; Youdim, M.B. Novel molecular targets of the neuroprotective/neurorescue multimodal iron chelating drug M30 in the mouse brain. Neuroscience 2011, 189, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Salkovic-Petrisic, M.; Knezovic, A.; Osmanovic-Barilar, J.; Smailovic, U.; Trkulja, V.; Riederer, P.; Amit, T.; Mandel, S.; Youdim, M.B. Multi-target iron-chelators improve memory loss in a rat model of sporadic Alzheimer’s disease. Life Sci. 2015, 136, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braymer, J.J.; Detoma, A.S.; Choi, J.S.; Ko, K.S.; Lim, M.H. Recent Development of Bifunctional Small Molecules to Study Metal-Amyloid-beta Species in Alzheimer’s Disease. Int. J. Alzheiers Dis. 2010, 2011, 623051. [Google Scholar]
- Moss, S.; Subramanian, V.; Acharya, K.R. High resolution crystal structure of substrate-free human neprilysin. J. Struct. Biol. 2018, 204, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Oefner, C.; Pierau, S.; Schulz, H.; Dale, G.E. Structural studies of a bifunctional inhibitor of neprilysin and DPP-IV. Acta Crystallogr. D 2007, 63, 975–981. [Google Scholar] [CrossRef]
- Oefner, C.; D’Arcy, A.; Hennig, M.; Winkler, F.K.; Dale, G.E. Structure of human neutral endopeptidase (Neprilysin) complexed with phosphoramidon. J. Mol. Biol. 2000, 296, 341–349. [Google Scholar] [CrossRef]
- Baranello, R.J.; Bharani, K.L.; Padmaraju, V.; Chopra, N.; Lahiri, D.K.; Greig, N.H.; Pappolla, M.A.; Sambamurti, K. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer’s disease. Curr. Alzheimers Res. 2015, 12, 32–46. [Google Scholar] [CrossRef] [Green Version]
- Hellstrom-Lindahl, E.; Ravid, R.; Nordberg, A. Age-dependent decline of neprilysin in Alzheimer’s disease and normal brain: Inverse correlation with Ab levels. Neurobiol. Aging 2008, 29, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Erdos, E.G.; Skidgel, R.A. Neutral endopeptidase 24.11 (enkephalinase) and related regulators of peptide hormones. FASEB J. 1989, 3, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Moss, S.; Subramanian, V.; Acharya, K.R. Crystal structure of peptide-bound neprilysin reveals key binding interactions. FEBS Lett. 2020, 594, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Feygina, E.E.; Katrukha, A.G.; Semenov, A.G. Neutral Endopeptidase (Neprilysin) in Therapy and Diagnostics: Yin and Yang. Biochemistry 2019, 84, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Bayes-Genis, A.; Barallat, J.; Richards, A.M. A Test in Context: Neprilysin: Function, Inhibition, and Biomarker. J. Am. Coll. Cardiol. 2016, 68, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.; Nalbantoglu, J.; Crine, P. Neutral endopeptidase can hydrolyze b-amyloid(1-40) but shows no effect on b-amyloid precursor protein metabolism. Peptides 1995, 16, 647–652. [Google Scholar] [CrossRef]
- Bayes-Genis, A.; Barallat, J.; Galan, A.; de Antonio, M.; Domingo, M.; Zamora, E.; Urrutia, A.; Lupon, J. Soluble neprilysin is predictive of cardiovascular death and heart failure hospitalization in heart failure patients. J. Am. Coll. Cardiol. 2015, 65, 657–665. [Google Scholar] [CrossRef]
- Carson, J.A.; Turner, A.J. b-amyloid catabolism: Roles for neprilysin (NEP) and other metallopeptidases? J. Neurochem. 2002, 81, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Hinton, D.J.; Choi, D.S. Protein kinase C-regulated abeta production and clearance. Int. J. Alzheimers Dis. 2011, 2011, 857368. [Google Scholar] [PubMed] [Green Version]
- Stock, A.J.; Kasus-Jacobi, A.; Pereira, H.A. The role of neutrophil granule proteins in neuroinflammation and Alzheimer’s disease. J. Neuroinflamm. 2018, 15, 240. [Google Scholar] [CrossRef]
- Kanemitsu, H.; Tomiyama, T.; Mori, H. Human neprilysin is capable of degrading amyloid beta peptide not only in the monomeric form but also the pathological oligomeric form. Neurosci. Lett. 2003, 350, 113–116. [Google Scholar] [CrossRef]
- Huang, S.M.; Mouri, A.; Kokubo, H.; Nakajima, R.; Suemoto, T.; Higuchi, M.; Staufenbiel, M.; Noda, Y.; Yamaguchi, H.; Nabeshima, T.; et al. Neprilysin-sensitive synapse-associated amyloid-beta peptide oligomers impair neuronal plasticity and cognitive function. J. Biol. Chem. 2006, 281, 17941–17951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, A.I. The metallobiology of Alzheimer’s disease. Trends Neurosci. 2003, 26, 207–214. [Google Scholar] [CrossRef]
- Huang, J.Y.; Hafez, D.M.; James, B.D.; Bennett, D.A.; Marr, R.A. Altered NEP2 expression and activity in mild cognitive impairment and Alzheimer’s disease. J. Alzheimers. Dis. 2012, 28, 433–441. [Google Scholar] [CrossRef]
- Wang, S.; Wang, R.; Chen, L.; Bennett, D.A.; Dickson, D.W.; Wang, D.S. Expression and functional profiling of neprilysin, insulin-degrading enzyme, and endothelin-converting enzyme in prospectively studied elderly and Alzheimer’s brain. J. Neurochem. 2010, 115, 47–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farris, W.; Schutz, S.G.; Cirrito, J.R.; Shankar, G.M.; Sun, X.; George, A.; Leissring, M.A.; Walsh, D.M.; Qiu, W.Q.; Holtzman, D.M.; et al. Loss of neprilysin function promotes amyloid plaque formation and causes cerebral amyloid angiopathy. Am. J. Pathol. 2007, 171, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, M.; Higuchi, M.; Takaki, Y.; Matsuba, Y.; Tanji, H.; Nemoto, M.; Tomita, N.; Matsui, T.; Iwata, N.; Mizukami, H.; et al. Cerebrospinal fluid neprilysin is reduced in prodromal Alzheimer’s disease. Ann. Neurol. 2005, 57, 832–842. [Google Scholar] [CrossRef]
- Rose, J.B.; Crews, L.; Rockenstein, E.; Adame, A.; Mante, M.; Hersh, L.B.; Gage, F.H.; Spencer, B.; Potkar, R.; Marr, R.A.; et al. Neuropeptide Y fragments derived from neprilysin processing are neuroprotective in a transgenic model of Alzheimer’s disease. J. Neurosci. 2009, 29, 1115–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, T.; Iwata, N.; Tsubuki, S.; Takaki, Y.; Takano, J.; Huang, S.M.; Suemoto, T.; Higuchi, M.; Saido, T.C. Somatostatin regulates brain amyloid b peptide Ab42 through modulation of proteolytic degradation. Nature Med. 2005, 11, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Iwata, N.; Mizukami, H.; Shirotani, K.; Takaki, Y.; Muramatsu, S.; Lu, B.; Gerard, N.P.; Gerard, C.; Ozawa, K.; Saido, T.C. Presynaptic localization of neprilysin contributes to efficient clearance of amyloid-beta peptide in mouse brain. J. Neurosci. 2004, 24, 991–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, N.; Sekiguchi, M.; Hattori, Y.; Takahashi, A.; Asai, M.; Ji, B.; Higuchi, M.; Staufenbiel, M.; Muramatsu, S.; Saido, T.C. Global brain delivery of neprilysin gene by intravascular administration of AAV vector in mice. Sci. Rep. 2013, 3, 1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, C.R.; Kemble, A.M.; Niewoehner, J.; Freskgard, P.O.; Urich, E. Brain shuttle neprilysin reduces central amyloid-b levels. PLoS ONE 2020, 15, e0229850. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, D.; Wang, Y.; Huang, H.; Zhao, Y.; Zhou, H. Meta-analysis of expression and function of neprilysin in Alzheimer’s disease. Neurosci. Lett. 2017, 657, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Dagliyan, O.; Dagyildiz, E.; Baris, I.; Kavakli, I.H.; Kizilel, S.; Turkay, M. Structure based discovery of small molecules to regulate the activity of human insulin degrading enzyme. PLoS ONE 2012, 7, e31787. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Di Muzio, E.; Ciaccio, C.; Sbardella, D.; Di Pierro, D.; Polticelli, F.; Coletta, M.; Marini, S. Multiple allosteric sites are involved in the modulation of insulin-degrading-enzyme activity by somatostatin. FEBS J. 2016, 283, 3755–3770. [Google Scholar] [CrossRef] [Green Version]
- Im, H.; Manolopoulou, M.; Malito, E.; Shen, Y.; Zhao, J.; Neant-Fery, M.; Sun, C.Y.; Meredith, S.C.; Sisodia, S.S.; Leissring, M.A.; et al. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J. Biol. Chem. 2007, 282, 25453–25463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malito, E.; Hulse, R.E.; Tang, W.J. Amyloid beta-degrading cryptidases: Insulin degrading enzyme, presequence peptidase, and neprilysin. Cell. Mol. Life Sci. 2008, 65, 2574–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Joachimiak, A.; Rosner, M.R.; Tang, W.J. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 2006, 443, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Kuo, W.L.; Yousef, M.; Rosner, M.R.; Tang, W.J. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem. Biophys. Res. Commun. 2006, 343, 1032–1037. [Google Scholar] [CrossRef]
- Kurochkin, I.V.; Guarnera, E.; Berezovsky, I.N. Insulin-degrading enzyme in the fight against Alzheimer’s disease. Trends Pharmacol. Sci. 2018, 39, 49–58. [Google Scholar] [CrossRef]
- Kurochkin, I.V.; Goto, S. Alzheimer’s b-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994, 345, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Amata, O.; Marino, T.; Russo, N.; Toscano, M. Human insulin-degrading enzyme working mechanism. J. Am. Chem. Soc. 2009, 131, 14804–14811. [Google Scholar] [CrossRef]
- Tundo, G.R.; Sbardella, D.; Ciaccio, C.; Grasso, G.; Gioia, M.; Coletta, A.; Polticelli, F.; Di Pierro, D.; Milardi, D.; Van Endert, P.; et al. Multiple functions of insulin-degrading enzyme: A metabolic crosslight? Crit. Rev. Biochem. Bol. Biol. 2017, 52, 554–582. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.Q.; Walsh, D.M.; Ye, Z.; Vekrellis, K.; Zhang, J.; Podlisny, M.B.; Rosner, M.R.; Safavi, A.; Hersh, L.B.; Selkoe, D.J. Insulin-degrading enzyme regulates extracellular levels of amyloid b-protein by degradation. J. Biol. Chem. 1998, 273, 32730–32738. [Google Scholar] [CrossRef] [Green Version]
- Perez, A.; Morelli, L.; Cresto, J.C.; Castano, E.M. Degradation of soluble amyloid b-peptides 1-40, 1-42, and the Dutch variant 1-40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem. Res. 2000, 25, 247–255. [Google Scholar] [CrossRef]
- Leal, M.C.; Magnani, N.; Villordo, S.; Buslje, C.M.; Evelson, P.; Castano, E.M.; Morelli, L. Transcriptional regulation of insulin-degrading enzyme modulates mitochondrial amyloid b (Ab) peptide catabolism and functionality. J. Biol. Chem. 2013, 288, 12920–12931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikanyika, N.L.; Parkington, H.C.; Smith, A.I.; Kuruppu, S. Powering amyloid b degrading enzymes: A possible therapy for Alzheimer’s disease. Neurochem. Res. 2019, 44, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- de Tullio, M.B.; Castelletto, V.; Hamley, I.W.; Martino Adami, P.V.; Morelli, L.; Castano, E.M. Proteolytically inactive insulin-degrading enzyme inhibits amyloid formation yielding non-neurotoxic Ab peptide aggregates. PLoS ONE 2013, 8, e59113. [Google Scholar] [CrossRef] [Green Version]
- Kurochkin, I.V. Insulin-degrading enzyme: Embarking on amyloid destruction. Trends Biochem. Sci. 2001, 26, 421–425. [Google Scholar] [CrossRef]
- Portelius, E.; Mattsson, N.; Pannee, J.; Zetterberg, H.; Gisslen, M.; Vanderstichele, H.; Gkanatsiou, E.; Crespi, G.A.; Parker, M.W.; Miles, L.A.; et al. Ex vivo 18O-labeling mass spectrometry identifies a peripheral amyloid b clearance pathway. Mol. Neurodegener. 2017, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Stargardt, A.; Gillis, J.; Kamphuis, W.; Wiemhoefer, A.; Kooijman, L.; Raspe, M.; Benckhuijsen, W.; Drijfhout, J.W.; Hol, E.M.; Reits, E. Reduced amyloid-b degradation in early Alzheimer’s disease but not in the APPswePS1dE9 and 3xTg-AD mouse models. Aging Cell 2013, 12, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xiang, Z.; Haroutunian, V.; Buxbaum, J.D.; Stetka, B.; Pasinetti, G.M. Insulin degrading enzyme activity selectively decreases in the hippocampal formation of cases at high risk to develop Alzheimer’s disease. Neurobiol. Aging 2007, 28, 824–830. [Google Scholar] [CrossRef]
- Miller, B.C.; Eckman, E.A.; Sambamurti, K.; Dobbs, N.; Chow, K.M.; Eckman, C.B.; Hersh, L.B.; Thiele, D.L. Amyloid-b peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 6221–6226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farris, W.; Mansourian, S.; Chang, Y.; Lindsley, L.; Eckman, E.A.; Frosch, M.P.; Eckman, C.B.; Tanzi, R.E.; Selkoe, D.J.; Guenette, S. Insulin-degrading enzyme regulates the levels of insulin, amyloid b-protein, and the b-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 4162–4167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, G.T.; Gonzalez, F.J.; Pahan, K. Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc. Natl. Acad. USA 2015, 112, 8445–8450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcello, E.; Borroni, B.; Pelucchi, S.; Gardoni, F.; Di Luca, M. ADAM10 as a therapeutic target for brain diseases: From developmental disorders to Alzheimer’s disease. Expert Opin. Ther. Targets 2017, 21, 1017–1026. [Google Scholar] [CrossRef]
- Huovila, A.P.; Turner, A.J.; Pelto-Huikko, M.; Karkkainen, I.; Ortiz, R.M. Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 2005, 30, 413–422. [Google Scholar] [CrossRef]
- Yuan, X.Z.; Sun, S.; Tan, C.C.; Yu, J.T.; Tan, L. The Role of ADAM10 in Alzheimer’s Disease. J. Alzheimers Dis. 2017, 58, 303–322. [Google Scholar] [CrossRef]
- Wolfsberg, T.G.; Primakoff, P.; Myles, D.G.; White, J.M. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: Multipotential functions in cell-cell and cell-matrix interactions. J. Cell. Biol. 1995, 131, 275–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzine, P.R.; Ettcheto, M.; Cano, A.; Busquets, O.; Marcello, E.; Pelucchi, S.; Di Luca, M.; Endres, K.; Olloquequi, J.; Camins, A.; et al. ADAM10 in Alzheimer’s disease: Pharmacological modulation by natural compounds and its role as a peripheral marker. Biomed. Pharmacother. 2019, 113, 108661. [Google Scholar] [CrossRef] [PubMed]
- Lammich, S.; Kamp, F.; Wagner, J.; Nuscher, B.; Zilow, S.; Ludwig, A.K.; Willem, M.; Haass, C. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5′-untranslated region. J. Biol. Chem. 2011, 286, 45063–45072. [Google Scholar] [CrossRef] [Green Version]
- Hartl, D.; May, P.; Gu, W.; Mayhaus, M.; Pichler, S.; Spaniol, C.; Glaab, E.; Bobbili, D.R.; Antony, P.; Koegelsberger, S.; et al. A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer disease. Mol. Psychiatry 2020, 25, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Endres, K.; Deller, T. Regulation of Alpha-Secretase ADAM10 In vitro and In vivo: Genetic, Epigenetic, and Protein-Based Mechanisms. Front. Mol. Neurosci. 2017, 10, 56. [Google Scholar] [CrossRef]
- Seipold, L.; Altmeppen, H.; Koudelka, T.; Tholey, A.; Kasparek, P.; Sedlacek, R.; Schweizer, M.; Bar, J.; Mikhaylova, M.; Glatzel, M.; et al. In vivo regulation of the A disintegrin and metalloproteinase 10 (ADAM10) by the tetraspanin 15. Cell Mol. Life Sci. 2018, 75, 3251–3267. [Google Scholar] [CrossRef]
- Lammich, S.; Kojro, E.; Postina, R.; Gilbert, S.; Pfeiffer, R.; Jasionowski, M.; Haass, C.; Fahrenholz, F. Constitutive and regulated a-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA 1999, 96, 3922–3927. [Google Scholar] [CrossRef] [Green Version]
- Seegar, T.C.M.; Killingsworth, L.B.; Saha, N.; Meyer, P.A.; Patra, D.; Zimmerman, B.; Janes, P.W.; Rubinstein, E.; Nikolov, D.B.; Skiniotis, G.; et al. Structural basis for regulated proteolysis by the a-Secretase ADAM10. Cell 2017, 171, 1638–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stawikowska, R.; Cudic, M.; Giulianotti, M.; Houghten, R.A.; Fields, G.B.; Minond, D. Activity of ADAM17 (a disintegrin and metalloprotease 17) is regulated by its noncatalytic domains and secondary structure of its substrates. J. Biol. Chem. 2013, 288, 22871–22879. [Google Scholar] [CrossRef] [Green Version]
- Tape, C.J.; Willems, S.H.; Dombernowsky, S.L.; Stanley, P.L.; Fogarasi, M.; Ouwehand, W.; McCafferty, J.; Murphy, G. Cross-domain inhibition of TACE ectodomain. Proc. Natl. Acad. Sci. USA 2011, 108, 5578–5583. [Google Scholar] [CrossRef] [Green Version]
- Janes, P.W.; Saha, N.; Barton, W.A.; Kolev, M.V.; Wimmer-Kleikamp, S.H.; Nievergall, E.; Blobel, C.P.; Himanen, J.P.; Lackmann, M.; Nikolov, D.B. Adam meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 2005, 123, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Postina, R.; Schroeder, A.; Dewachter, I.; Bohl, J.; Schmitt, U.; Kojro, E.; Prinzen, C.; Endres, K.; Hiemke, C.; Blessing, M.; et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Investig. 2004, 113, 1456–1464. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Q.; Qu, D.H.; Wang, K. Therapeutic approaches to Alzheimer’s disease through stimulating of non-amyloidogenic processing of amyloid precursor protein. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2389–2403. [Google Scholar]
- Marcello, E.; Saraceno, C.; Musardo, S.; Vara, H.; de la Fuente, A.G.; Pelucchi, S.; Di Marino, D.; Borroni, B.; Tramontano, A.; Perez-Otano, I.; et al. Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer’s disease. J. Clin. Investig. 2013, 123, 2523–2538. [Google Scholar] [CrossRef] [PubMed]
- Peron, R.; Vatanabe, I.P.; Manzine, P.R.; Camins, A.; Cominetti, M.R. Alpha-Secretase ADAM10 Regulation: Insights into Alzheimer’s Disease Treatment. Pharmaceuticals 2018, 11, 12. [Google Scholar] [CrossRef] [Green Version]
- Prox, J.; Bernreuther, C.; Altmeppen, H.; Grendel, J.; Glatzel, M.; D’Hooge, R.; Stroobants, S.; Ahmed, T.; Balschun, D.; Willem, M.; et al. Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J. Neurosci. 2013, 33, 12915–12928. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, M.; Liu, Y.; Yu, J.; Yang, H.; Fan, D.; Chui, D. Copper downregulates neprilysin activity through modulation of neprilysin degradation. J. Alzheimers Dis. 2010, 19, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Lang, M.; Fan, Q.; Wang, L.; Zheng, Y.; Xiao, G.; Wang, X.; Wang, W.; Zhong, Y.; Zhou, B. Inhibition of human high-affinity copper importer Ctr1 orthologous in the nervous system of drosophila ameliorates Ab42-induced Alzheimer’s disease-like symptoms. Neurobiol. Aging 2013, 34, 2604–2612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mital, M.; Bal, W.; Fraczyk, T.; Drew, S.C. Interplay between copper, neprilysin, and N-truncation of b-amyloid. Inorg. Chem. 2018, 57, 6193–6197. [Google Scholar] [CrossRef]
- Banerjee, P.; Sahoo, A.; Anand, S.; Bir, A.; Chakrabarti, S. The oral iron chelator, deferasirox, reverses the age-dependent alterations in iron and amyloid-b homeostasis in rat brain: Implications in the therapy of Alzheimer’s disease. J. Alzheimers Dis. 2016, 49, 681–693. [Google Scholar] [CrossRef]
- Grasso, G.; Pietropaolo, A.; Spoto, G.; Pappalardo, G.; Tundo, G.R.; Ciaccio, C.; Coletta, M.; Rizzarelli, E. Copper(I) and copper(II) inhibit Ab peptides proteolysis by insulin-degrading enzyme differently: Implications for metallostasis alteration in Alzheimer’s disease. Chem. Eur. J. 2011, 17, 2752–2762. [Google Scholar] [CrossRef] [Green Version]
- Bellia, F.; Lanza, V.; Ahmed, I.M.M.; Garcia-Vinuales, S.; Veiss, E.; Arizzi, M.; Calcagno, D.; Milardi, D.; Grasso, G. Site directed mutagenesis of insulin-degrading enzyme allows singling out the molecular basis of peptidase versus E1-like activity: The role of metal ions. Metallomics 2019, 11, 278–281. [Google Scholar] [CrossRef]
- Kitazawa, M.; Cheng, D.; Laferla, F.M. Chronic copper exposure exacerbates both amyloid and tau pathology and selectively dysregulates cdk5 in a mouse model of AD. J. Neurochem. 2009, 108, 1550–1560. [Google Scholar] [CrossRef] [Green Version]
- Maras, J.S.; Das, S.; Sharma, S.; Sukriti, S.; Kumar, J.; Vyas, A.K.; Kumar, D.; Bhat, A.; Yadav, G.; Choudhary, M.C.; et al. Iron-Overload triggers ADAM-17 mediated inflammation in Severe Alcoholic Hepatitis. Sci. Rep. 2018, 8, 10264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.H.; Yoo, Y.M. Altered APP Carboxyl-Terminal Processing Under Ferrous Iron Treatment in PC12 Cells. Korean J. Physiol. Pharmacol. 2013, 17, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.H.; Zhang, W.; Gao, H.L.; Zhong, M.L.; Huang, T.T.; Guo, R.F.; Liu, N.N.; Li, D.D.; Li, Y.; et al. Copper chelators promote nonamyloidogenic processing of AbPP via MT1/2 /CREB-dependent signaling pathways in AbPP/PS1 transgenic mice. J. Pineal Res. 2018, 65, e12502. [Google Scholar] [CrossRef] [PubMed]




| Chemical Agents | Clinical Trials (Periods) | Status |
|---|---|---|
| PBT-2 | Phase 2 (December, 2006-December, 2007) | Completed (NCT00471211) |
| Phase 2 (September, 2011-January, 2014) | Completed (ACTRN12611001008910) | |
| Phase 2 (July, 2013-January, 2015) | Completed (ACTRN12613000777796) | |
| Deferiprone (DFP) | Phase 2 (October, 2016-September, 2019) | Completed (NCT02728843) |
| Phase 2 (November, 2017-October, 2019) | Completed (ACTRN12617001578392) | |
| Phase 2 (January, 2018-December, 2021 (Estimated)] | Recruiting (NCT03234686) |
| Chemical Agents | Animal Models | Outcomes | References |
|---|---|---|---|
| Clioquinol (CQ) | C. elegans | Enhancement of neuroprotective effect | [142] |
| DP-109 | Transgenic mouse | Extension of life span and Reduction of amyloid plaques | [152,153] |
| DP-460 | Transgenic mouse | Extension of life span and Reduction of amyloid plaques | [152,153] |
| Rat | Detrimental effect on learning | [154] | |
| D-Penicillamine | Transgenic mouse | Improvement of cognitive ability | [163] |
| Tetrathiomolybdate | Transgenic mouse | Decrease of the inflammatory cytokines | [167] |
| Chemical Agents | Animal Model | Outcomes | References |
|---|---|---|---|
| Deferoxamine (DFO) | Transgenic mouse | Improvement of cognitive function | [280] |
| Mouse | Improvement of memory | [281] | |
| Rat | Improvement of memory | [282] | |
| Deferiprone (DFP) | Transgenic mouse | Improvement of cognitive function | [285,286,287] |
| M30 | Rat | Recovery of memory impairment | [292,293] |
| Mouse | Increase of neuroprotective effect Improvement of memory | [294] | |
| HLA20 | Rat | Recovery of memory impairment | [292,293] |
| NEP | IDE | ADAM10 | ||
|---|---|---|---|---|
| Cu(I/II) | Expression Levels | Decreased | Need to study | Increased |
| Activity | Decreased | Decreased | Need to study | |
| Fe(II/III) | Expression Levels | Need to study | Need to study | Increased |
| Activity | Decreased | Need to study | Need to study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Lee, H.J. Redox-Active Metal Ions and Amyloid-Degrading Enzymes in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 7697. https://doi.org/10.3390/ijms22147697
Kim N, Lee HJ. Redox-Active Metal Ions and Amyloid-Degrading Enzymes in Alzheimer’s Disease. International Journal of Molecular Sciences. 2021; 22(14):7697. https://doi.org/10.3390/ijms22147697
Chicago/Turabian StyleKim, Namdoo, and Hyuck Jin Lee. 2021. "Redox-Active Metal Ions and Amyloid-Degrading Enzymes in Alzheimer’s Disease" International Journal of Molecular Sciences 22, no. 14: 7697. https://doi.org/10.3390/ijms22147697