Transcriptomics Reveals Fast Changes in Salicylate and Jasmonate Signaling Pathways in Shoots of Carbonate-Tolerant Arabidopsis thaliana under Bicarbonate Exposure
Abstract
:1. Introduction
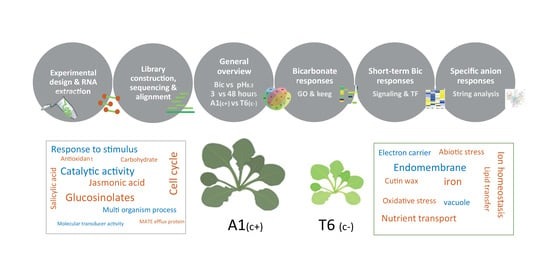
2. Results and Discussion
2.1. Fitness Characterization of A. thaliana Plants under High pH and Bicarbonate Stress
2.2. Nutrient Content Profile
2.3. Analysis of RNA-Seq Data
2.4. Gene Ontology (GO) and Kioto Encyclopedia of Genes and Genomes (KEGG) Pathways under Bicarbonate Stress
2.5. Signal Perception and Signal Transduction
2.5.1. Receptor-Like Kinases and Receptor-Like Proteins (RLK/RLP)
2.5.2. Peroxidases
2.5.3. Glutathione-Related
2.5.4. Calmodulin
2.5.5. Transcription Factors
2.6. Protein–Protein Interaction Network Functional Enrichment Analysis with Specific Bic-Induced Genes
2.7. Mechanisms Underlying Differential bic Toxicity and Tolerance in A. thaliana Demes
3. Materials and Methods
3.1. Plant Material
3.2. Growth Conditions
3.3. Physiological Trait Measurements
3.4. RNA Isolation, Library Construction, and Sequencing
3.5. Gene Ontology, KEGG Pathway, and Functional Protein Association Network Analysis of DEGs
3.6. Relative Expression Analysis
3.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ANOVA | Analysis of variance |
| A. thaliana | Arabidopsis thaliana |
| cDNA | Complementary deoxyribonucleic acid |
| BTP | 1,3-Bis[tris(hydroxymethyl)methylamino]propane |
| DEG | Differentially expressed genes |
| FDR | Fold discovery rate |
| GO | Gene Ontology |
| GS | Glucosinolate |
| GST | Gluthatione-S-transferase |
| ICP-OES | Inductively coupled plasma atomic emission spectroscopy |
| JA | Jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LFC | Log2 fold change |
| MATE | Multidrug and Toxic Compound Extrusion |
| MES | 2-(N-Morpholino)ethanesulfonic acid hydrate, 4-morpholineethanesulfonic acid |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NCBI | National Center for Biotechnology Information |
| Adj p-value | Adjusted p-value |
| RLK | Receptor-like kinase |
| RLP | Receptor-like protein |
| RNA | Ribonucleic acid |
| ROS | Reactive oxygen species |
| qPCR | Quantitative Polymerase Chain Reaction |
| TF | Transcription factor |
| SA | Salicylic acid |
References
- Al-Busaidi, A.S.; Cookson, P. Salinity–pH Relationships in Calcareous Soils. J. Agric. Mar. Sci. 2003, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Loeppert, R.H.; Suarez, D.L. Carbonate and Gypsum. In Methods of Soil Analysis. Part 3. Chemical Methods; Sparks, D.L., Page, A.L., Hemke, P.A., Loeppert, R.H., Soltanpour, M.A., Tabatabai, M.A., Johnston, C.T., Summer, M.E., Eds.; Soil Science Society of America: Madison, WI, USA; American Society of Agronomy: Lincoln, NE, USA, 1996; pp. 437–474. ISBN 9780891188254. [Google Scholar]
- Hu, H.; Rappel, W.-J.; Occhipinti, R.; Ries, A.; Böhmer, M.; You, L.; Xiao, C.; Engineer, C.B.; Boron, W.F.; Schroeder, J.I. Distinct Cellular Locations of Carbonic Anhydrases Mediate Carbon Dioxide Control of Stomatal Movements. Plant Physiol. 2015, 169, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Aboukila, E.F.; Nassar, I.N.; Rashad, M.; Hafez, M.; Norton, J.B. Reclamation of calcareous soil and improvement of squash growth using brewers’ spent grain and compost. J. Saudi Soc. Agric. Sci. 2018, 17, 390–397. [Google Scholar] [CrossRef] [Green Version]
- Morrissey, J.; Guerinot, M.L. Iron Uptake and Transport in Plants: The Good, the Bad, and the Ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T. Understanding the Complexity of Iron Sensing and Signaling Cascades in Plants. Plant Cell Physiol. 2019, 60, 1440–1446. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [Green Version]
- López-Millán, A.-F.; Grusak, M.A.; Abadía, A.; Abadía, J. Iron deficiency in plants: An insight from proteomic approaches. Front. Plant Sci. 2013, 4, 254. [Google Scholar] [CrossRef] [Green Version]
- Donnini, S.; De Nisi, P.; Gabotti, D.; Tato, L.; Zocchi, G. Adaptive strategies of Parietaria diffusa (M.&K.) to calcareous habitat with limited iron availability. Plant Cell Environ. 2012, 35, 1171–1184. [Google Scholar] [CrossRef]
- Higuchi, K.; Ono, K.; Araki, S.; Nakamura, S.; Uesugi, T.; Makishima, T.; Ikari, A.; Hanaoka, T.; Sue, M. Elongation of barley roots in high-pH nutrient solution is supported by both cell proliferation and differentiation in the root apex. Plant Cell Environ. 2017, 40, 1609–1617. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.-H.; Liu, W.-C.; Zhang, X.-W.; Lu, Y.-T. Ethylene Inhibits Root Elongation during Alkaline Stress through AUXIN1 and Associated Changes in Auxin Accumulation. Plant Physiol. 2015, 168, 1777–1791. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Bai, X.; Chen, C.; Chen, Q.; Cai, H.; Li, Y.; Ji, W.; Zhai, H.; Lv, D.; Luo, X.; et al. GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 2011, 77, 285–297. [Google Scholar] [CrossRef]
- Lucena, C.; Romera, F.J.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene participates in the regulation of Fe deficiency responses in strategy I plants and in rice. Front. Plant Sci. 2015, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Öztürk, M.; Sharma, S.; Gucel, S. Effect of sodium carbonate-induced salinity–alkalinity on some key osmoprotectants, protein profile, antioxidant enzymes, and lipid peroxidation in two mulberry (Morus alba L.) cultivars. J. Plant Interact. 2014, 9, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, J.; Jiang, W.; Liu, J.; Yang, S.; Gai, J.; Li, Y. Identification and Analysis of NaHCO3 Stress Responsive Genes in Wild Soybean (Glycine soja) Roots by RNA-seq. Front. Plant Sci. 2016, 7, 1842. [Google Scholar] [CrossRef] [Green Version]
- Duan, X.; Yu, Y.; Zhang, Y.; Chen, C.; Duanmu, H.; Cao, L.; Sun, M.; Sun, X.; Zhu, Y. A potential efflux boron transporter gene GsBOR2, positively regulates Arabidopsis bicarbonate tolerance. Plant Sci. 2018, 274, 284–292. [Google Scholar] [CrossRef]
- Ge, Y.; Li, Y.; Zhu, Y.-M.; Bai, X.; Lv, D.; Guo, D.; Ji, W.; Cai, H. Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 2010, 10, 153. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Li, S.; Tian, S.; Wang, B.; Zhao, X. Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba). PLoS ONE. 2017, 12, 0185732. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Gao, C.; Wang, L.; Zheng, L.; Yang, C.; Wang, Y. Comprehensive transcriptional profiling of NaHCO3-stressed Tamarix hispida roots reveals networks of responsive genes. Plant Mol. Biol. 2004, 84, 145–157. [Google Scholar] [CrossRef]
- Gong, B.; Zhang, C.; Li, X.; Wen, D.; Wang, S.; Shi, Q.; Wang, X. Identification of NaCl and NaHCO3 stress responsive proteins in tomato roots using iTRAQ-based analysis. Biochem. Biophys Res. Commun. 2014, 446, 417–422. [Google Scholar] [CrossRef]
- Waters, B.M.; Amundsen, K.; Graef, G. Gene Expression Profiling of Iron Deficiency Chlorosis Sensitive and Tolerant Soybean Indicates Key Roles for Phenylpropanoids under Alkalinity Stress. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jieng, C.J.; Liang, Z.W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef]
- Terés, J.; Busoms, S.; Pérez-Martín, L.; Luís-Villaroya, A.; Flis, P.; Álvarez-Fernández, A.; Tolrà, R.; Salt, D.E.; Poschenrieder, C. Soil carbonate drives local adaptation in Arabidopsis thaliana. Plant Cell Environ. 2019, 42, 2384–2398. [Google Scholar] [CrossRef]
- Chen, L.; Wang, G.; Chen, P.; Zhu, H.; Wang, S.; Ding, Y. Shoot-Root Communication Plays a Key Role in Physiological Alterations of Rice (Oryza sativa) Under Iron Deficiency. Front. Plant Sci. 2018, 9, 757. [Google Scholar] [CrossRef]
- García, M.J.; Corpas, F.J.; Lucena, C.; Alcántara, E.; Pérez-Vicente, R.; Zamarreño, A.M.; Bacaicoa, E.; García-Mina, J.M.; Bauer, P.; Romera, F.J. A Shoot Fe Signaling Pathway Requiring the OPT3 Transporter Controls GSNO Reductase and Ethylene in Arabidopsis thaliana Roots. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Tudela-Isanta, M.; Ladouceur, E.; Wijayasinghe, M.; Pritchard, H.W.; Mondoni, A. The seed germination niche limits the distribution of some plant species in calcareous or siliceous alpine bedrocks. Alp. Bot. 2018, 128, 83–95. [Google Scholar] [CrossRef]
- Lin, J.; Li, X.; Zhang, Z.; Yu, X.; Gao, Z.; Wang, Y.; Wang, J.; Li, Z.; Mu, C. Salinity-alkalinity tolerance in wheat: Seed germination, early seedling growth, ion relations and solute accumulation. African J. Agric. Res. 2012, 7, 467–474. [Google Scholar] [CrossRef]
- Meng, C.; Quan, T.-Y.; Li, Z.-Y.; Cui, K.L.; Liang, Y.; Dai, J.L.; Xia, G.M.; Liu, S.W. Transcriptome profiling reveals the genetic basis of alkalinity tolerance in wheat. BMC Genomics. 2017, 18, 24. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, E.J.; Waters, B.M. Alkaline stress and iron deficiency regulate iron uptake and riboflavin synthesis gene expression differently in root and leaf tissue: Implications for iron deficiency chlorosis. J. Exp. Bot. 2016, 67, 5671–5685. [Google Scholar] [CrossRef]
- Bergmann, W. Ernährungsstörungen bei Kulturpflanzen, 3rd ed.; Gustav Fischer Verlag: Jena, Germany, 1993; ISBN 13: 978-3-334-60414-4. [Google Scholar]
- Sisó-Terraza, P.; Luis-Villarroya, A.; Fourcroy, P.; Briat, J.F.; Abadía, A.; Gaymard, F.; Abadía, J.; Álvarez-Fernández, A. Accumulation and Secretion of Coumarinolignans and other Coumarins in Arabidopsis thaliana Roots in Response to Iron Deficiency at High pH. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Yang, H.; Lü, X.; Wu, P.H.; Liang, Z.; Ooi, M.K.J. Does high pH give a reliable assessment of the effect of alkaline soil on seed germination? A case study with Leymus chinensis (Poaceae). Plant Soil. 2015, 394, 35–43. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Schmidt, W. A Digital Compendium of Genes Mediating the Reversible Phosphorylation of Proteins in Fe-Deficient Arabidopsis Roots. Front. Plant Sci. 2013, 4, 173. [Google Scholar] [CrossRef] [Green Version]
- Steidele, C.E.; Stam, R. Multi-omics approach highlights differences between functional RLP classes in Arabidopsis thaliana. Biorxiv 2020. [Google Scholar] [CrossRef]
- Sakai, T.; Van Der Honing, H.; Nishioka, M.; Uehara, Y.; Takahashi, M.; Fujisawa, N.; Saji, N.; Seki, M.; Shinozaki, K.; Jones, M.A.; et al. Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 2008, 53, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Tarutani, Y.; Sasaki, A.; Yasuda, M.; Nakashita, H.; Yoshida, S.; Yamaguchi, I.; Suzuki, Y. Identification of Three Clones Which Commonly Interact with the Kinase Domains of Highly Homologous Two Receptor-Like Kinases, RLK902 and RKL1. Biosci. Biotechnol. Biochem. 2004, 68, 2581–2587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, H.; Fan, X.; Sugimoto, K.; Fukao, Y.; Peng, L.; Shikanai, T. CHLORORESPIRATORY REDUCTION 9 is a Novel Factor Required for Formation of Subcomplex A of the Chloroplast NADH Dehydrogenase-Like Complex. Plant Cell Physiol. 2016, 57, 2122–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Msilini, N.; Oueslati, S.; Amdouni Chebbi, M.; Ksouri, R.; Lachaâl, M.; Ouerghi, Z. Variability of phenolic content and antioxidant activity of two lettuce varieties under Fe deficiency. J. Sci. Food Agric. 2013, 93, 2016–2021. [Google Scholar] [CrossRef]
- Ranieri, A.; Castagna, A.; Baldan, B.; Soldatini, G.F. Iron deficiency differently affects peroxidase isoforms in sunflower. J. Exp. Bot. 2001, 52, 25–35. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Tokunaga, N.; Kaneta, T.; Sato, S.; Sato, Y. Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiol. Plant 2009, 136, 237–249. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-Wide Identification of Arabidopsis LBD29 Target Genes Reveals the Molecular Events behind Auxin-Induced Cell Reprogramming during Callus Formation. Plant Cell Physiol. 2018, 59, 749–760. [Google Scholar] [CrossRef]
- Fiorucci, A.-S.; Bourbousse, C.; Concia, L.; Rougée, M.; Deton-Cabanillas, A.-F.; Zabulon, G.; Layat, E.; Latrasse, D.; Kim, S.K.; Chaumont, N.; et al. Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination. Genome Biol. 2019, 20, 100. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.S.; Ozgur, R.; Uzilday, B.; Turkan, I.; Roriz, M.; Rangel, A.; Carvalho, S.M.P.; Vasconcelos, M.W. Understanding the Role of the Antioxidant System and the Tetrapyrrole Cycle in Iron Deficiency Chlorosis. Plants 2019, 8, 348. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Milla, M.A.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36602–36615. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 871, 1836. [Google Scholar] [CrossRef] [Green Version]
- Piślewska-Bednarek, M.; Nakano, R.T.; Hiruma, K.; Pastorczyk, M.; Sanchez-Vallet, A.; Singkravanit-Ogawa, S.; Ciesiolka, D.; Takano, Y.; Molina, P.; Schulze-Lefert, P.; et al. Glutathione Transferase U13 Functions in Pathogen-Triggered Glucosinolate Metabolism. Plant Physiol. 2018, 176, 538–551. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Yang, Y.; Liu, K.; Zhang, L.; Gou, H.; Sun, T.; Wang, H. Involvement of endogenous salicylic acid in iron-deficiency responses in Arabidopsis. J. Exp. Bot. 2016, 67, 4179–4193. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhang, X.; Yang, H.; Agerbirk, N.; Qiu, Y.; Wang, H.; Shen, D.; Song, J.; Li, X. Aromatic Glucosinolate Biosynthesis Pathway in Barbarea vulgaris and its Response to Plutella xylostella Infestation. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Czerniawski, P.; Bednarek, P. Glutathione S-Transferases in the Biosynthesis of Sulfur-Containing Secondary Metabolites in Brassicaceae Plants. Front. Plant Sci. 2018, 9, 1639. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Selective Binding of Glutathione Conjugates of Fatty Acid Derivatives by Plant Glutathione Transferases. J. Biol. Chem. 2009, 284, 21249–21256. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Chen, X.; Lin, W.; Chen, S.; Lu, D.; Niu, Y.; Li, L.; Cheng, C.; McCormack, M.; Sheen, J.; et al. Bifurcation of Arabidopsis, N.L.R. Immune Signaling via Ca2+-Dependent Protein Kinases. PLoS Pathog. 2013, 9, 1003127. [Google Scholar] [CrossRef] [Green Version]
- Scholz, S.S.; Vadassery, J.; Heyer, M.; Reichelt, M.; Snedden, W.A.; Boland, W.; Mithöfer, A. Mutation of the Arabidopsis Calmodulin-Like Protein CML37 Deregulates the Jasmonate Pathway and Enhances Susceptibility to Herbivory. Mol. Plant 2014, 7, 1712–1726. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Cheval, C.; Laohavisit, A.; Hocking, B.; Chiasson, D.; Olson, T.; Shirasu, K.; Faulkner, C.; Gilliham, M. A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017, 215, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Chai, M.; Yang, J.; Ning, G.; Wang, G.; Ma, H. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 Gene Is Required for Male Fertility through Regulating Callose Metabolism during Microsporogenesis. Plant Physiol. 2014, 164, 1893–1904. [Google Scholar] [CrossRef] [Green Version]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Parra, E.; López-Matas, M.A.; Fründt, C.; Gutierrez, C. Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell. 2004, 16, 2350–2363. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.A.; LaCroix, I.S.; Gerber, S.A.; Guerinot, M.L. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. USA 2019, 116, 24933–24942. [Google Scholar] [CrossRef] [Green Version]
- Kroh, G.E.; Pilon, M. Connecting the negatives and positives of plant iron homeostasis. New Phytol. 2019, 223, 1052–1055. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, B.; Bauer, P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, C.; Gaillochet, C.; Medzihradszky, A.; Busch, W.; Daum, G.; Krebs, M.; Kehle, A.; Lohmann, J.U. A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 2014, 28, 438–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, O.S.; Davies, K.A.; Chang, J.; Adrian, J.; Rowe, M.H.; Ballenger, C.E.; Bergmann, D.C. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 2014, 345, 1605–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haga, N.; Kobayashi, K.; Suzuki, T.; Maeo, K.; Kubo, M.; Ontani, M.; Mitsuda, N.; Demura, T.; Nakamura, K.; Jürgens, G.; et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011, 157, 706–717. [Google Scholar] [CrossRef] [Green Version]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef] [Green Version]
- Finatto, T.; Viana, V.E.; Woyann, L.G.; Busanello, C.; da Maia, L.C.; de Oliveira, A.C. Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol Biol. 2018, 41, 533–544. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004, 16, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Grunewald, W.; De Smet, I.; Lewis, D.R.; Löfke, C.; Jansen, L.; Goeminne, G.; Vanden-Bossche, R.; Karimi, M.; De Rybel, B.; Vanholme, B.; et al. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 1554–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, T.; Uemura, T.; Nemoto, K.; Daito, M.; Nozawa, A.; Sawasaki, T.; Arimura, G. Tyrosine kinase-dependent defense responses against herbivory in Arabidopsis. Front. Plant Sci. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Bahieldin, A.; Atef, A.; Edris, S.; Gadalla, N.O.; Ramadan, A.M.; Hassan, S.M.; Al Attas, S.G.; Al-Kordy, M.; Al-Hajar, A.S.M.; Sabir, J.; et al. Multifunctional activities of ERF109 as affected by salt stress in Arabidopsis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raines, T.; Shanks, C.; Cheng, C.Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Vanková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef]
- Liu, W.; Li, Q.; Wang, Y.; Wu, T.; Yang, Y.; Zhang, X.; Han, Z.; Xu, X. Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem. Biophys. Res. Commun. 2017, 491, 862–868. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, Z.; Cao, G.; Li, J.; Wei, J.; Tsuge, T.; Gu, H.; Aoyama, T.; Qu, L.J. TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance. Mol. Plant 2014, 7, 601–615. [Google Scholar] [CrossRef] [Green Version]
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N.V. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2011, 75, 107–127. [Google Scholar] [CrossRef]
- Saelim, L.; Akiyoshi, N.; Tan, T.T.; Ihara, A.; Yamaguchi, M.; Hirano, K.; Matsuoka, M.; Demura, T.; Ohtani, M. Arabidopsis Group IIId ERF proteins positively regulate primary cell wall-type CESA genes. J. Plant Res. 2019, 132, 117–129. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Tian, H.; Wang, S.; Chen, J.G. The small ethylene response factor ERF96 is involved in the regulation of the abscisic acid response in arabidopsis. Front. Plant Sci. 2015, 6, 1064. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Park, J.H.; Kim, J.; Kim, J.J.; Sunghyun, H.; Kim, J.; Kim, J.H.; Woo, H.R.; Hyeron, C.; Lim, P.O.; et al. Time-evolving genetic networks reveal a nac troika that negatively regulates leaf senescence in arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 4930–4939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naranjo-Arcos, M.A.; Maurer, F.; Meiser, J.; Pateyron, S.; Fink-Straube, C.; Bauer, P. Dissection of iron signaling and iron accumulation by overexpression of subgroup Ib bHLH039 protein. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivitz, A.B.; Hermand, V.; Curie, C.; Vert, G. Arabidopsis bHLH100 and bHLH101 Control Iron Homeostasis via a FIT-Independent Pathway. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Geu-Flores, F.; Moldrup, M.E.; Böttcher, C.; Olsen, C.E.; Scheel, D.; Halkier, B.A. Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in arabidopsis. Plant Cell. 2011, 23, 2456–2469. [Google Scholar] [CrossRef] [Green Version]
- Jeschke, V.; Weber, K.; Moore, S.S.; Burow, M. Coordination of Glucosinolate Biosynthesis and Turnover Under Different Nutrient Conditions. Front. Plant Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Howarth, J.R.; Parmar, S.; Barraclough, P.B.; Hawkesford, M.J. A sulphur deficiency-induced gene, sdi1, involved in the utilization of stored sulphate pools under sulphur-limiting conditions has potential as a diagnostic indicator of sulphur nutritional status. Plant Biotechnol. J. 2009, 7, 200–209. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Altmann, M.; Alkofer, A.; Epple, P.M.; Dangl, J.L.; Falter-Braun, P. LSU network hubs integrate abiotic and biotic stress responses via interaction with the superoxide dismutase FSD2. J. Exp. Bot. 2017, 68, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Kawaguchi, R.; Morikawa-Ichinose, T.; Allahham, A.; Kim, S.J.; Maruyama-Nakashita, A. Sulfur deficiency-induced glucosinolate catabolism attributed to two β-glucosidases, bglu28 and bglu30, is required for plant growth maintenance under sulfur deficiency. Plant Cell Physiol. 2020, 61, 803–813. [Google Scholar] [CrossRef]
- Clark, T.J.; Lu, Y. Analysis of loss-of-function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate-derived amino acid contents. Plant Physiol. 2015, 168, 1512–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Schutter, K.; Joubès, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuck, E.; Van der Schueren, E.; Beeckman, T.; Kushnir, S.; Inzé, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell. 2007, 19, 211–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisa, M.; Huang, Y.; Benhamed, M.; Raynaud, C. The plant DNA damage response: Signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, T.; Ito, M.; Sumikura, T.; Nakayama, A. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014, 78, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Wei, J.; Zhao, Y.; Yan, H.; Sun, B.; Huang, J.; Wang, Q. Glucose signaling positively regulates aliphatic glucosinolate biosynthesis. J. Exp. Bot. 2013, 64, 1097–1109. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, L.; Bartoli, C.G.; Lamattina, L. Glutathione and ascorbic acid protect Arabidopsis plants against detrimental effects of iron deficiency. J. Exp. Bot. 2013, 64, 3169–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; He, Z.; Pandey, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 2002, 277, 5360–5368. [Google Scholar] [CrossRef] [Green Version]
- Fukudome, A.; Aksoy, E.; Wu, X.; Kumar, K.; Jeong, I.S.; May, K.; Russell, W.K.; Koiwa, H. Arabidopsis CPL4 is an essential C-terminal domain phosphatase that suppresses xenobiotic stress responses. Plant J. 2014, 80, 27–39. [Google Scholar] [CrossRef]
- Lucena, C.; Romera, F.J.; Rojas, C.L.; García, M.J.; Alcántara, E.; Pérez-Vicente, R. Bicarbonate blocks the expression of several genes involved in the physiological responses to Fe deficiency of Strategy I plants. Funct Plant Biol. 2007, 34, 1002–1009. [Google Scholar] [CrossRef]
- Wang, N.; Dong, X.; Chen, Y.; Ma, B.; Yao, C.; Ma, F.; Liu, Z. Direct and bicarbonate-induced iron deficiency differently affect iron translocation in Kiwifruit roots. Plants 2020, 9, 1578. [Google Scholar] [CrossRef]
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; ISBN 9780124735439. [Google Scholar]
- Poschenrieder, C.; Fernández, J.A.; Rubio, L.; Pérez-Martín, L.; Terés, J.; Barceló, J. Transport and use of bicarbonate in plants: Current knowledge and challenges ahead. Int. J. Mol. Sci. 2018, 19, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busoms, S.; Teres, J.; Huang, X.-Y.; Bomblies, K.; Danku, J.; Douglas, A.; Weigel, D.; Poschenrieder, C.; Salt, D.E. Salinity Is an Agent of Divergent Selection Driving Local Adaptation of Arabidopsis to Coastal Habitats. Plant Physiol. 2015, 168, 915–929. [Google Scholar] [CrossRef] [PubMed]
- Scholl, R.L.; May, S.T.; Ware, D.H. Seed and Molecular Resources for Arabidopsis. Plant Physiol. 2000, 124, 1477–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Yekutieli, D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Stat. 2011, 29, 1165–1188. [Google Scholar]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, 122–129. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huertas-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Dekkers, B.J.W.; Willems, L.; Bassel, G.W.; van Bolderern-Veldkamp, R.P.M.; Ligterink, W.; Hilhorst, H.W.M.; Bentsink, L. Identification of Reference Genes for RT–qPCR Expression Analysis in Arabidopsis and Tomato Seeds. Plant Cell Physiol. 2012, 53, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Martín, L.; Busoms, S.; Tolrà, R.; Poschenrieder, C. Transcriptomics Reveals Fast Changes in Salicylate and Jasmonate Signaling Pathways in Shoots of Carbonate-Tolerant Arabidopsis thaliana under Bicarbonate Exposure. Int. J. Mol. Sci. 2021, 22, 1226. https://doi.org/10.3390/ijms22031226
Pérez-Martín L, Busoms S, Tolrà R, Poschenrieder C. Transcriptomics Reveals Fast Changes in Salicylate and Jasmonate Signaling Pathways in Shoots of Carbonate-Tolerant Arabidopsis thaliana under Bicarbonate Exposure. International Journal of Molecular Sciences. 2021; 22(3):1226. https://doi.org/10.3390/ijms22031226
Chicago/Turabian StylePérez-Martín, Laura, Silvia Busoms, Roser Tolrà, and Charlotte Poschenrieder. 2021. "Transcriptomics Reveals Fast Changes in Salicylate and Jasmonate Signaling Pathways in Shoots of Carbonate-Tolerant Arabidopsis thaliana under Bicarbonate Exposure" International Journal of Molecular Sciences 22, no. 3: 1226. https://doi.org/10.3390/ijms22031226