The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms
Abstract
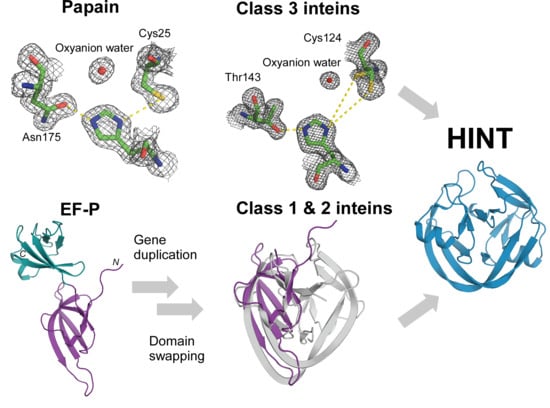
:1. Introduction
2. Results
2.1. Self-Cleavage Activity and Inhibition of Class 3 Inteins by Protease Inhibitors
2.2. Conversion of a Class 1 Intein into a Class 3 Intein
2.3. The Active Site of the MchDnaB1 Class 3 Intein
2.4. Molecular Dynamics Simulation
2.5. The Catalytic Mechanism of Class 3 Inteins
3. Discussion
4. Methods
4.1. Cloning of Class 3 Intein Expression Vectors
4.2. Expression and Purification of MchDnaB1_HN, MchDnaB1_HAA, and gp41-1_WCT
4.3. Proteolytic Inhibition Assays
4.4. Protein Cis-Splicing Tests
4.5. Crystallization and Structure Determination of MchDnaB1_HN, MchDnaB1_HAA, gp41-1_WCT
4.6. Molecular Dynamics Simulation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BIL | Bacterial Intein-Like |
| HINT | Hedgehog/INTein |
| Hh-C | the C-terminal domain of the Hedgehog protein or hog protein |
| IMAC | immobilized metal affinity chromatography |
| IPTG | isopropyl-β-D-thiogalactoside |
| MchDnaB1 intein | DnaB1 intein from Mycobacterium chimaera |
| PDB | Protein Data Bank |
| r.m.s.d. | root-mean-square deviation |
| HSQC | heteronuclear single quantum correlation |
| PEG | polyethylene glycol |
| PMSF | phenylmethanesulfonyl fluoride |
| DTT | dithiothreitol. |
References
- Hirata, R.; Ohsumk, Y.; Nakano, A.; Kawasaki, H.; Suzuki, K.; Anraku, Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 6726–6733. [Google Scholar] [PubMed]
- Paulus, H. Protein Splicing and Related Forms of Protein Autoprocessing. Annu. Rev. Biochem. 2000, 69, 447–496. [Google Scholar] [CrossRef] [PubMed]
- Novikova, O.; Topilina, N.; Belfort, M. Enigmatic Distribution, Evolution, and Function of Inteins. J. Biol. Chem. 2014, 289, 14490–14497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaï, H.; Mikula, K.M.; Oeemig, J.S.; Zhou, D.; Li, M.; Wlodawer, A. Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins. J. Mol. Biol. 2017, 429, 3942–3956. [Google Scholar] [CrossRef] [PubMed]
- Novikova, O.; Jayachandran, P.; Kelley, D.S.; Morton, Z.; Merwin, S.; Topilina, N.I.; Belfort, M. Intein Clustering Suggests Functional Importance in Different Domains of Life. Mol. Biol. Evol. 2016, 33, 783–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perler, F.B. InBase: The Intein Database. Nucleic Acids Res. 2002, 30, 383–384. [Google Scholar] [CrossRef] [Green Version]
- Pietrokovski, S. Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 1994, 3, 2340–2350. [Google Scholar] [CrossRef] [Green Version]
- Noren, C.; Wang, J.; Perler, F. Dissecting the Chemistry of Protein Splicing and Its Applications. Angew. Chem. Int. Ed. Engl. 2000, 39, 450–466. [Google Scholar] [CrossRef]
- Tori, K.; Dassa, B.; Johnson, M.A.; Southworth, M.W.; Brace, L.E.; Ishino, Y.; Pietrokovski, S.; Perler, F.B. Splicing of the mycobacteriophage Bethlehem DnaB intein: Identification of a new mechanistic class of inteins that contain an obligate block F nucleophile. J. Biol. Chem. 2010, 285, 2515–2526. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.M.; Porter, J.A.; Young, K.E.; Koonin, E.V.; Beachy, P.A.; Leahy, D.J. Crystal Structure of a Hedgehog Autoprocessing Domain: Homology between Hedgehog and Self-Splicing Proteins. Cell 1997, 91, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Amitai, G.; Belenkiy, O.; Dassa, B.; Shainskaya, A.; Pietrokovski, S. Distribution and function of new bacterial intein-like protein domains. Mol. Microbiol. 2002, 47, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Southworth, M.W.; Benner, J.; Perler, F.B. An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. EMBO J. 2000, 19, 5019–5026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brace, L.E.; Southworth, M.W.; Tori, K.; Cushing, M.L.; Perler, F.B. The Deinococcus radiodurans Snf2 intein caught in the act: Detection of the Class 3 intein signature Block F branched intermediate. Protein Sci. 2010, 19, 1525–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tori, K.; Perler, F.B. Expanding the Definition of Class 3 Inteins and Their Proposed Phage Origin. J. Bacteriol. 2011, 193, 2035–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranko, A.S.; Oeemig, J.S.; Zhou, N.; Kajander, T.; Wlodawer, A.; Iwai, H. Structure-based engineering and comparison of novel split inteins for protein ligation. Mol. BioSyst. 2014, 10, 1023–1034. [Google Scholar] [CrossRef]
- Aranko, A.S.; Oeemig, J.S.; Iwai, H. Structural basis for protein trans-splicing by a bacterial intein-like domain—Protein ligation without nucleophilic side chains. FEBS J. 2013, 280, 3256–3269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Southworth, M.W.; Herrmann, T.; Brace, L.; Perler, F.B.; Wüthrich, K. NMR structure of a KlbA intein precursor from Methanococcus jannaschii. Protein Sci. 2007, 16, 1316–1328. [Google Scholar] [CrossRef] [Green Version]
- Aranko, A.S.; Oeemig, J.S.; Kajander, T.; Iwai, H. Intermolecular domain swapping induces intein-mediated protein alternative splicing. Nat. Chem. Biol. 2013, 9, 616–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Benner, J.; Perler, F.B. Protein Splicing in the Absence of an Intein Penultimate Histidine. J. Biol. Chem. 2000, 275, 20431–20435. [Google Scholar] [CrossRef] [Green Version]
- Tori, K.; Cheriyan, M.; Pedamallu, C.S.; Contreras, M.A.; Perler, F.B. The Thermococcus kodakaraensis Tko CDC21-1 Intein Activates Its N-Terminal Splice Junction in the Absence of a Conserved Histidine by a Compensatory Mechanism. Biochemistry 2012, 51, 2496–2505. [Google Scholar] [CrossRef]
- Kelley, D.S.; Lennon, C.W.; Li, Z.; Miller, M.R.; Banavali, N.K.; Li, H.; Belfort, M. Mycobacterial DnaB helicase intein as oxidative stress sensor. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Aranko, A.; Wlodawer, A.; Iwaï, H. Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 2014, 27, 263–271. [Google Scholar] [CrossRef] [Green Version]
- Botos, I.; Wlodawer, A. The expanding diversity of serine hydrolases. Curr. Opin. Struct. Biol. 2007, 17, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Mills, K.V.; Johnson, M.A.; Perler, F.B. Protein Splicing: How Inteins Escape from Precursor Proteins. J. Biol. Chem. 2014, 289, 14498–14505. [Google Scholar] [CrossRef] [Green Version]
- Turini, P.; Kurooka, S.; Steer, M.; Corbascio, A.N.; Singer, T.P. The action of phenylmethylsulfonyl fluoride on human acetylcholinesterase, chymotyrpsin and trypsin. J. Pharmacol. Exp. Ther. 1969, 167, 98–104. [Google Scholar]
- Borutaite, V.; Brown, G.C. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Lett. 2001, 500, 114–118. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Abraham, M.H.; Zissimos, A.M. Fast Calculation of van der Waals Volume as a Sum of Atomic and Bond Contributions and Its Application to Drug Compounds. J. Org. Chem. 2003, 68, 7368–7373. [Google Scholar] [CrossRef]
- Ciragan, A.; Aranko, A.S.; Tascon, I.; Iwai, H. Salt-inducible Protein Splicing in cis and trans by Inteins from Extremely Halophilic Archaea as a Novel Protein-Engineering Tool. J. Mol. Biol. 2016, 428, 4573–4588. [Google Scholar] [CrossRef] [PubMed]
- Southworth, M.; Yin, J.; Perler, F.B. Rescue of protein splicing activity from a Magnetospirillum magnetotacticum intein-like element. Biochem. Soc. Trans. 2004, 32, 250–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, H.M.; Mikula, K.M.; Li, M.; Wlodawer, A.; Iwaï, H. The crystal structure of the naturally split gp41-1 intein guides the engineering of orthogonal split inteins from cis-splicing inteins. FEBS J. 2019, 287, 1886–1898. [Google Scholar] [CrossRef]
- Mizutani, R.; Nogami, S.; Kawasaki, M.; Ohya, Y.; Anraku, Y.; Satow, Y. Protein-splicing Reaction via a Thiazolidine Intermediate: Crystal Structure of the VMA1-derived Endonuclease Bearing the N and C-terminal Propeptides. J. Mol. Biol. 2002, 316, 919–929. [Google Scholar] [CrossRef]
- Poland, B.W.; Xu, M.-Q.; Quiocho, F.A. Structural Insights into the Protein Splicing Mechanism of PI-SceI. J. Biol. Chem. 2000, 275, 16408–16413. [Google Scholar] [CrossRef] [Green Version]
- Oeemig, J.S.; Zhou, D.; Kajander, T.; Wlodawer, A.; Iwaï, H. NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein. J. Mol. Biol. 2012, 421, 85–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wierenga, R.K. The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 2001, 492, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Buller, A.R.; Townsend, C.A. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc. Natl. Acad. Sci. USA 2013, 110, E653–E661. [Google Scholar] [CrossRef] [Green Version]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- McLachlan, A. Gene duplications in the structural evolution of chymotrypsin. J. Mol. Biol. 1979, 128, 49–79. [Google Scholar] [CrossRef]
- Beyer, H.M.; Mikula, K.M.; Kudling, T.V.; Iwai, H. Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site. Extremophiles 2019, 23, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Morihara, K.; Oka, T. α-Chymotrypsin as the catalyst for peptide synthesis. Biochem. J. 1977, 163, 531–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, H.; Hart, S.A.; Schink, A.; Pollok, B.A. Sortase-Mediated Protein Ligation: A New Method for Protein Engineering. J. Am. Chem. Soc. 2004, 126, 2670–2671. [Google Scholar] [CrossRef]
- Peat, T.S.; Newman, J.; Waldo, G.S.; Berendzen, J.; Terwilliger, T.C. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 å resolution. Structure 1998, 6, 1207–1214. [Google Scholar] [CrossRef] [Green Version]
- Tori, K.; Perler, F.B. The Arthrobacter Species FB24 Arth_1007 (DnaB) Intein Is a Pseudogene. PLoS ONE 2011, 6, e26361. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laakso, L.M. Dali server update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef]
- Teng, Y.-B.; Ma, X.-X.; He, Y.-X.; Jiang, Y.-L.; Du, J.; Xiang, C.; Chen, Y.; Zhou, C.-Z. Crystal structure of Arabidopsis translation initiation factor eIF-5A2. Proteins Struct. Funct. Bioinform. 2009, 77, 736–740. [Google Scholar] [CrossRef]
- Hanawa-Suetsugu, K.; Sekine, S.-I.; Sakai, H.; Hori-Takemoto, C.; Terada, T.; Unzai, S.; Tame, J.R.H.; Kuramitsu, S.; Shirouzu, M.; Yokoyama, S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. USA 2004, 101, 9595–9600. [Google Scholar] [CrossRef] [Green Version]
- Dodson, G. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Gherardini, P.F.; Wass, M.N.; Helmer-Citterich, M.; Sternberg, M.J.E. Convergent Evolution of Enzyme Active Sites Is not a Rare Phenomenon. J. Mol. Biol. 2007, 372, 817–845. [Google Scholar] [CrossRef]
- Tomii, K.; Sawada, Y.; Honda, S. Convergent evolution in structural elements of proteins investigated using cross profile analysis. BMC Bioinform. 2012, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W. H. Freeman: New York, NY, USA, 2015. [Google Scholar]
- Ellilä, S.; Jurvansuu, J.M.; Iwai, H. Evaluation and comparison of protein splicing by exogenous inteins with foreign exteins inEscherichia coli. FEBS Lett. 2011, 585, 3471–3477. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, F.; Ciragan, A.; Iwaï, H. Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 2015, 116, 42–49. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.-W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Svensson, O.; Gilski, M.; Nurizzo, D.; Bowler, M.W. Multi-position data collection and dynamic beam sizing: Recent improvements to the automatic data-collection algorithms on MASSIF-1. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 433–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, O.; Monaco, S.; Popov, A.N.; Nurizzo, D.; Bowler, M.W. The fully automatic characterization and data collection from crystals of biological macromolecules. Acta Cryst. 2015, D71, 1757–1767. [Google Scholar] [CrossRef]
- Bowler, M.W.; Nurizzo, D.; Barrett, R.; Beteva, A.; Bodin, M.; Caserotto, H.; Delagenière, S.; Dobias, F.; Flot, D.; Giraud, T.; et al. MASSIF-1: A beamline dedicated to the fully automatic characterisation and data collection from crystals of biological macromolecules. J. Synchrotron Radiat. 2015, 22, 1540–1547. [Google Scholar] [CrossRef]
- Langer, G.G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef]
- Joosten, R.P.; Salzemann, J.; Bloch, V.; Stockinger, H.; Berglund, A.-C.; Blanchet, C.; Bongcam-Rudloff, E.; Combet, C.; Da Costa, A.L.; Deleage, G.; et al. PDB_REDO: Automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009, 42, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.A.; York, D.M.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beyer, H.M.; Virtanen, S.I.; Aranko, A.S.; Mikula, K.M.; Lountos, G.T.; Wlodawer, A.; Ollila, O.H.S.; Iwaï, H. The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms. Int. J. Mol. Sci. 2020, 21, 8367. https://doi.org/10.3390/ijms21218367
Beyer HM, Virtanen SI, Aranko AS, Mikula KM, Lountos GT, Wlodawer A, Ollila OHS, Iwaï H. The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms. International Journal of Molecular Sciences. 2020; 21(21):8367. https://doi.org/10.3390/ijms21218367
Chicago/Turabian StyleBeyer, Hannes M., Salla I. Virtanen, A. Sesilja Aranko, Kornelia M. Mikula, George T. Lountos, Alexander Wlodawer, O. H. Samuli Ollila, and Hideo Iwaï. 2020. "The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms" International Journal of Molecular Sciences 21, no. 21: 8367. https://doi.org/10.3390/ijms21218367