A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis
Abstract
:1. Introduction
2. Results
2.1. Design of Immunogenic KRAS Peptides and Production of KRAS Antibodies
2.2. Detection of KRAS in Transfected HEK-293 Cells and Electroporated Mouse Skeletal Muscles
2.3. Detection of Unprenylated and Prenylated Forms of KRAS in Mouse Tissues
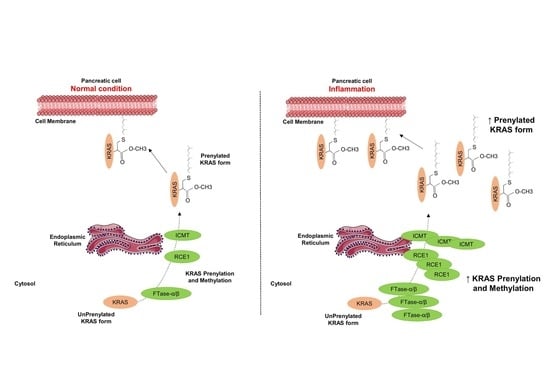
2.4. Inflammation Promotes KRAS Prenylation
2.5. Pancreas Inflammation Promotes the Expression of KRAS-Prenylating Enzymes
2.6. KRAS Prenylation and Expression of KRAS-Prenylating Enzymes Correlate with the Susceptibility of the Pancreas to Initiate Carcinogenesis
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Antibody Production
4.3. Enzyme-Linked Immunosorbent Assay (ELISA)
4.4. Cell Culture, Plasmid Transfection, and Pharmacological Treatments
4.5. Ex Vivo Culture of Dissociated Mouse Pancreas
4.6. Skeletal Muscle Electroporation
4.7. Fluorescence-Activated Cell Sorting (FACS) and Real-Time Quantitative PCR (RTqPCR)
4.8. Western Blotting
4.9. Immunolabeling and Quantification
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malumbres, M.; Barbacid, M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer 2003, 3, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Oltean, S.; Bates, D.O. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, F.D.; Lopes, M.S.; Zhou, M.; Court, H.; Ponce, O.; Fiordalisi, J.J.; Gierut, J.J.; Cox, A.D.; Haigis, K.M.; Philips, M.R. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. USA 2015, 112, 779–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussinov, R.; Tsai, C.J.; Chakrabarti, M.; Jang, H. A New View of Ras Isoforms in Cancers. Cancer Res. 2016, 76, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [Green Version]
- Hingorani, S.R.; Petricoin, E.F.; Maitra, A.; Rajapakse, V.; King, C.; Jacobetz, M.A.; Ross, S.; Conrads, T.P.; Veenstra, T.D.; Hitt, B.A.; et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003, 4, 437–450. [Google Scholar] [CrossRef] [Green Version]
- Chavan, T.S.; Muratcioglu, S.; Marszalek, R.; Jang, H.; Keskin, O.; Gursoy, A.; Nussinov, R.; Gaponenko, V. Plasma membrane regulates Ras signaling networks. Cell Logist 2015, 5, e1136374. [Google Scholar] [CrossRef] [Green Version]
- Papke, B.; Der, C.J. Drugging RAS: Know the enemy. Science 2017, 355, 1158–1163. [Google Scholar] [CrossRef] [Green Version]
- Palsuledesai, C.C.; Distefano, M.D. Protein prenylation: Enzymes, therapeutics, and biotechnology applications. ACS Chem. Biol. 2015, 10, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Berglund, L.; Bjorling, E.; Oksvold, P.; Fagerberg, L.; Asplund, A.; Szigyarto, C.A.; Persson, A.; Ottosson, J.; Wernerus, H.; Nilsson, P.; et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteom. 2008, 7, 2019–2027. [Google Scholar] [CrossRef] [Green Version]
- Voskuil, J.L. The challenges with the validation of research antibodies. F1000Res 2017, 6, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, A.M.; Ozkan-Dagliyan, I.; Vaseva, A.V.; Fer, N.; Strathern, L.A.; Hobbs, G.A.; Tessier-Cloutier, B.; Gillette, W.K.; Bagni, R.; Whiteley, G.R.; et al. Evaluation of the selectivity and sensitivity of isoform- and mutation-specific RAS antibodies. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmick, M.; Vartak, N.; Papke, B.; Kovacevic, M.; Truxius, D.C.; Rossmannek, L.; Bastiaens, P.I.H. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 2014, 157, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndt, N.; Sebti, S.M. Measurement of protein farnesylation and geranylgeranylation in vitro, in cultured cells and in biopsies, and the effects of prenyl transferase inhibitors. Nat. Protoc 2011, 6, 1775–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, C.; Schuhmacher, A.J.; Canamero, M.; Grippo, P.J.; Verdaguer, L.; Perez-Gallego, L.; Dubus, P.; Sandgren, E.P.; Barbacid, M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007, 11, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Qu, C.; Konieczny, S.F. Pancreatic Acinar Cell 3-Dimensional Culture. Biol. Protoc. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Lubeseder-Martellato, C. Isolation, Culture and Differentiation of Primary Acinar Epithelial Explants from Adult Murine Pancreas. Biol. Protoc. 2013, 3. [Google Scholar] [CrossRef]
- Frohner, I.E.; Mudrak, I.; Schuchner, S.; Anrather, D.; Hartl, M.; Sontag, J.M.; Sontag, E.; Wadzinski, B.E.; Preglej, T.; Ellmeier, W.; et al. PP2AC Phospho-Tyr(307) Antibodies Are Not Specific for this Modification but Are Sensitive to Other PP2AC Modifications Including Leu(309) Methylation. Cell Rep. 2020, 30, 3171–3182. [Google Scholar] [CrossRef] [Green Version]
- Frohner, I.E.; Mudrak, I.; Kronlachner, S.; Schuchner, S.; Ogris, E. Antibodies recognizing the C terminus of PP2A catalytic subunit are unsuitable for evaluating PP2A activity and holoenzyme composition. Sci Signal. 2020, 13. [Google Scholar] [CrossRef]
- Schuchner, S.; Behm, C.; Mudrak, I.; Ogris, E. The Myc tag monoclonal antibody 9E10 displays highly variable epitope recognition dependent on neighboring sequence context. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef]
- Kazi, A.; Xiang, S.; Yang, H.; Chen, L.; Kennedy, P.; Ayaz, M.; Fletcher, S.; Cummings, C.; Lawrence, H.R.; Beato, F.; et al. Dual Farnesyl and Geranylgeranyl Transferase Inhibitor Thwarts Mutant KRAS-Driven Patient-Derived Pancreatic Tumors. Clin. Cancer Res. 2019, 25, 5984–5996. [Google Scholar] [CrossRef] [PubMed]
- Diver, M.M.; Pedi, L.; Koide, A.; Koide, S.; Long, S.B. Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT. Nature 2018, 553, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, I.; Hampton, S.E.; Ashall, L.; Hildebrandt, E.R.; Kutlik, R.A.; Manandhar, S.P.; Floyd, B.J.; Smith, H.E.; Dozier, J.K.; Distefano, M.D.; et al. 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg. Med. Chem. 2016, 24, 160–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampton, S.E.; Dore, T.M.; Schmidt, W.K. Rce1: Mechanism and inhibition. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 157–174. [Google Scholar] [CrossRef]
- Steinhart, Z.; Pavlovic, Z.; Chandrashekhar, M.; Hart, T.; Wang, X.; Zhang, X.; Robitaille, M.; Brown, K.R.; Jaksani, S.; Overmeer, R.; et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med. 2017, 23, 60–68. [Google Scholar] [CrossRef]
- Desai, B.M.; Oliver-Krasinski, J.; De Leon, D.D.; Farzad, C.; Hong, N.; Leach, S.D.; Stoffers, D.A. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet beta cell, regeneration. J. Clin. Investig. 2007, 117, 971–977. [Google Scholar] [CrossRef] [Green Version]
- Schakman, O.; Gilson, H.; de Coninck, V.; Lause, P.; Verniers, J.; Havaux, X.; Ketelslegers, J.M.; Thissen, J.P. Insulin-like growth factor-I gene transfer by electroporation prevents skeletal muscle atrophy in glucocorticoid-treated rats. Endocrinology 2005, 146, 1789–1797. [Google Scholar] [CrossRef] [Green Version]
- Assi, M.; Dauguet, N.; Jacquemin, P. DIE-RNA: A Reproducible Strategy for the Digestion of Normal and Injured Pancreas, Isolation of Pancreatic Cells from Genetically Engineered Mouse Models and Extraction of High Quality RNA. Front. Physiol. 2018, 9, 129. [Google Scholar] [CrossRef] [Green Version]







| Applications | Rabbit #5 | Rabbit #6 | Rabbit #7 | Rabbit #8 | |
|---|---|---|---|---|---|
| Peptide-1 | Western blot | K (+++); N (+) | K (+++); N (+++); H (++) | K (+++); N (+) | K (+++); N (+++) |
| Cell labeling | K (+++); N (+) | K (+++); N (++) | K (+); N (++) | K (+++); N (+++) | |
| Tissue labeling | K (+++); N (+) | K(+++); N (++) | None | K (+++); N (+++) | |
| Applications | Rabbit #30 | Rabbit #31 | Rabbit #32 | Rabbit #33 | |
| Peptide-2 | Western blot | K (+) | K (+++); N (+++); H (++) | None | None |
| Cell labeling | K (+) | K (++) | None | None | |
| Tissue labeling | None | None | None | None |
| Gene | Forward (5′–3′) | Reverse (5′–3′) | Species |
|---|---|---|---|
| FTase-α | ATGGACGACGGGTTTCTGAG | TAAAGGCTCGTTCGCTCCTC | Mus musculus |
| FTase-β | TCCCCTGTTTGGTCAGAACC | GCATCCAGACACTCATAGGCA | Mus musculus |
| Rce1 | GTGTCCTGGTAGTGTCCAGC | CAGGACAACCTTCAGCCCAT | Mus musculus |
| Icmt | CCGCCGGCTCTTCCG | AGCCAAGGAAACAAGCTCTGA | Mus musculus |
| Rpl04 | CGCAACATCCCTGGTATTACT | TGTGCATGGGCAGGTTATAGT | Mus musculus |
| Antibody | Reference | Dilution | Antibody Incubation Condition |
|---|---|---|---|
| FTase-α | Sc-373749, SCBT | 1/200 | 5% milk, overnight, 4 °C |
| FTase-β | Sc-46664, SCBT | 1/200 | 5% milk, overnight, 4 °C |
| RCE1 | Ab62531, Abcam | 1/500 | 5% milk, overnight, 4 °C |
| ICMT | HPA032025-100UL, Sigma Aldrich | 1/500 | 5% milk, overnight, 4 °C |
| RAS (EP1125Y) | Ab52939, Abcam | 1/1000 | 5% milk, overnight, 4 °C |
| KRAS | WH0003845M1, Sigma Aldrich | 1/500 | 5% milk, overnight, 4 °C |
| KRAS | OP24-100UG, Merck Chemicals | 1/1000 | 5% milk, overnight, 4 °C |
| KRAS | 415700, Thermo Fisher Scientific | 1/1000 | 5% milk, overnight, 4 °C |
| Homemade KRAS antibodies | 1/250 | 5% milk or BSA, overnight, 4 °C |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Assi, M.; Pirlot, B.; Stroobant, V.; Thissen, J.-P.; Jacquemin, P. A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 6361. https://doi.org/10.3390/ijms21176361
Assi M, Pirlot B, Stroobant V, Thissen J-P, Jacquemin P. A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis. International Journal of Molecular Sciences. 2020; 21(17):6361. https://doi.org/10.3390/ijms21176361
Chicago/Turabian StyleAssi, Mohamad, Boris Pirlot, Vincent Stroobant, Jean-Paul Thissen, and Patrick Jacquemin. 2020. "A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis" International Journal of Molecular Sciences 21, no. 17: 6361. https://doi.org/10.3390/ijms21176361