Non-Coding RNA: Role in Gestational Diabetes Pathophysiology and Complications
Abstract
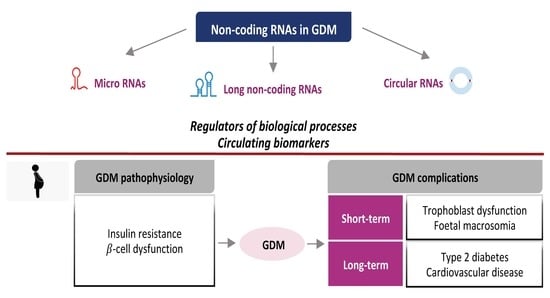
:1. Introduction and GDM Overview
2. General Aspects of NcRNAs
3. NcRNAs: Insights into the Pathophysiology of GDM
3.1. Insulin Resistance and Metabolic Adaptations
3.2. β-Cell Dysfunction
4. NcRNAs as Candidate Circulating Biomarkers of GDM
| Study | Groups | Stage of Pregnancy | Source | miRNA |
|---|---|---|---|---|
| Cao et al. [98] | 85 GDM | 16–28 weeks | Plasma | miR-16-5p, miR-17-5p, miR-20a-5p (↑) |
| 72 CTRL | ||||
| Lamadrid-Romero et al. [107] | 13 GDM | 1st trimester | Serum | 1st trimester: |
| 12 CTRL | miR-125b-5p, miR-183-5p, miR-200b-3p, miR-1290 (↑) | |||
| 24 GDM | 2nd trimester | 2nd trimester: | ||
| 24 CTRL | miR-183-5p, miR-128-5p (↑) | |||
| miR-125b-5p in the (↓) | ||||
| 20 GDM | 3rd trimester | 3rd trimester: | ||
| miR-137 (↑) | ||||
| miR-183-5p, miR-200b-3p (↓) | ||||
| Wander et al. [99] | 36 GDM | 7–23 weeks | Plasma | miR-155-5p, miR-21-3p, miR-210-3p, miR-146b-5p, miR-223-3p, miR-517-5p, miR-29a-3p (↑) |
| 80 CTRL | ||||
| Zhu et al. [97] | 10 GDM | 16–19 weeks | Plasma | miR-16-5p, miR-17-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p (↑) |
| 10 CTRL | ||||
| Tagoma et al. [101] | 13 GDM | 25–27 weeks | Plasma | miR-195-5p, miR-222-3p (↑) |
| 9 CTRL | ||||
| Sebastiani et al. [92] | 21 GDM | 24–33 weeks | Plasma | miR-330-3p (↑) |
| 10 CTRL | ||||
| Zhao et al. [104] | 24 GDM | 16–19 weeks | Serum | miR-29a, miR-123, miR-222 (↓) |
| 24 CTRL | ||||
| Pheiffer et al [105] | 53 GDM | 26–27 weeks | Serum | miR-222-3p, miR-20a-5p (↓) |
| 28 CTRL | ||||
| Yoffe et al. [106] | 23 GDM | 9–11 weeks | Plasma | miR-223, miR-23a (↑) |
| 20 CTRL |
5. NcRNAs and GDM Complications
5.1. Short-Term Complications
5.2. Long-Term Complications
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care 2019, 42, S13–S28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casagrande, S.S.; Linder, B.; Cowie, C.C. Prevalence of gestational diabetes and subsequent Type 2 diabetes among U.S. women. Diabetes Res. Clin. Pract. 2018, 141, 200–208. [Google Scholar] [CrossRef] [PubMed]
- HAPO Study Cooperative Research Group; Metzger, B.E.; Lowe, L.P.; Dyer, A.R.; Trimble, E.R.; Chaovarindr, U.; Coustan, D.R.; Hadden, D.R.; McCance, D.R.; Hod, M.; et al. Hyperglycemia and adverse pregnancy outcomes. N. Engl. J. Med. 2008, 358, 1991–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzger, B.E.; Persson, B.; Lowe, L.P.; Dyer, A.R.; Cruickshank, J.K.; Deerochanawong, C.; Halliday, H.L.; Hennis, A.J.; Liley, H.; Ng, P.C.; et al. Hyperglycemia and adverse pregnancy outcome study: Neonatal glycemia. Pediatrics 2010, 126, e1545–e1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szmuilowicz, E.D.; Josefson, J.L.; Metzger, B.E. Gestational Diabetes Mellitus. Endocrinol. Metab. Clin. North Am. 2019, 48, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.D.; Mathiesen, E.R.; Hansen, T.; Pedersen, O.; Jensen, D.M.; Lauenborg, J.; Damm, P. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: The role of intrauterine hyperglycemia. Diabetes Care 2008, 31, 340–346. [Google Scholar] [CrossRef] [Green Version]
- D.A.I. Study Group. The DAI prospective study on macrovascular complications in patients with type 2 diabetes. Characteristics of the study population. Ann. Ist. Super Sanita 2001, 37, 289–296. [Google Scholar]
- Yu, Y.; Arah, O.A.; Liew, Z.; Cnattingius, S.; Olsen, J.; Sorensen, H.T.; Qin, G.; Li, J. Maternal diabetes during pregnancy and early onset of cardiovascular disease in offspring: Population based cohort study with 40 years of follow-up. BMJ 2019, 367, l6398. [Google Scholar] [CrossRef] [Green Version]
- Clausen, T.D.; Mathiesen, E.R.; Hansen, T.; Pedersen, O.; Jensen, D.M.; Lauenborg, J.; Schmidt, L.; Damm, P. Overweight and the metabolic syndrome in adult offspring of women with diet-treated gestational diabetes mellitus or type 1 diabetes. J. Clin. Endocrinol. Metab. 2009, 94, 2464–2470. [Google Scholar] [CrossRef] [Green Version]
- International Association of Diabetes; Pregnancy Study Groups Consensus Panel; Metzger, B.E.; Gabbe, S.G.; Persson, B.; Buchanan, T.A.; Catalano, P.A.; Damm, P.; Dyer, A.R.; Leiva, A.; et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010, 33, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Pu, J.; Zhao, B.; Wang, E.J.; Nimbal, V.; Osmundson, S.; Kunz, L.; Popat, R.A.; Chung, S.; Palaniappan, L.P. Racial/Ethnic Differences in Gestational Diabetes Prevalence and Contribution of Common Risk Factors. Paediatr. Perinat. Epidemiol. 2015, 29, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, N.; Nachum, Z.; Green, M.S. The prevalence of gestational diabetes mellitus recurrence-effect of ethnicity and parity: A metaanalysis. Am. J. Obstet. Gynecol. 2015, 213, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Li, X.; Chen, R.; Zhou, X.; Liu, C.; Wu, J.; Xu, S.; Wang, W.; Xiao, M.; Xiong, G.; et al. Greater early and mid-pregnancy gestational weight gain are associated with increased risk of gestational diabetes mellitus: A prospective cohort study. Clin. Nutr. ESPEN 2017, 22, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Filardi, T.; Panimolle, F.; Crescioli, C.; Lenzi, A.; Morano, S. Gestational Diabetes Mellitus: The Impact of Carbohydrate Quality in Diet. Nutrients 2019, 11, 1549. [Google Scholar] [CrossRef] [Green Version]
- Filardi, T.; Panimolle, F.; Lenzi, A.; Morano, S. Bisphenol A and Phthalates in Diet: An Emerging Link with Pregnancy Complications. Nutrients 2020, 12, 525. [Google Scholar] [CrossRef] [Green Version]
- Filardi, T.; Tavaglione, F.; Di Stasio, M.; Fazio, V.; Lenzi, A.; Morano, S. Impact of risk factors for gestational diabetes (GDM) on pregnancy outcomes in women with GDM. J. Endocrinol. Investig. 2018, 41, 671–676. [Google Scholar] [CrossRef]
- Pintaudi, B.; Fresa, R.; Dalfra, M.; Dodesini, A.R.; Vitacolonna, E.; Tumminia, A.; Sciacca, L.; Lencioni, C.; Marcone, T.; Lucisano, G.; et al. The risk stratification of adverse neonatal outcomes in women with gestational diabetes (STRONG) study. Acta Diabetol. 2018, 55, 1261–1273. [Google Scholar] [CrossRef]
- Di Cianni, G.; Miccoli, R.; Volpe, L.; Lencioni, C.; Del Prato, S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab. Res. Rev. 2003, 19, 259–270. [Google Scholar] [CrossRef]
- Santangelo, C.; Zicari, A.; Mandosi, E.; Scazzocchio, B.; Mari, E.; Morano, S.; Masella, R. Could gestational diabetes mellitus be managed through dietary bioactive compounds? Current knowledge and future perspectives. Br. J. Nutr. 2016, 115, 1129–1144. [Google Scholar] [CrossRef]
- Plows, J.F.; Stanley, J.L.; Baker, P.N.; Reynolds, C.M.; Vickers, M.H. The Pathophysiology of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 3342. [Google Scholar] [CrossRef] [Green Version]
- Butler, A.E.; Cao-Minh, L.; Galasso, R.; Rizza, R.A.; Corradin, A.; Cobelli, C.; Butler, P.C. Adaptive changes in pancreatic beta cell fractional area and beta cell turnover in human pregnancy. Diabetologia 2010, 53, 2167–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalano, P.M. Trying to understand gestational diabetes. Diabet Med. 2014, 31, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Barbour, L.A.; McCurdy, C.E.; Hernandez, T.L.; Kirwan, J.P.; Catalano, P.M.; Friedman, J.E. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care 2007, 30 (Suppl. 2), S112–S119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santangelo, C.; Filardi, T.; Perrone, G.; Mariani, M.; Mari, E.; Scazzocchio, B.; Masella, R.; Brunelli, R.; Lenzi, A.; Zicari, A.; et al. Cross-talk between fetal membranes and visceral adipose tissue involves HMGB1-RAGE and VIP-VPAC2 pathways in human gestational diabetes mellitus. Acta Diabetol. 2019, 56, 681–689. [Google Scholar] [CrossRef]
- Lekva, T.; Norwitz, E.R.; Aukrust, P.; Ueland, T. Impact of Systemic Inflammation on the Progression of Gestational Diabetes Mellitus. Curr. Diab. Rep. 2016, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef] [Green Version]
- Jayabalan, N.; Lai, A.; Ormazabal, V.; Adam, S.; Guanzon, D.; Palma, C.; Scholz-Romero, K.; Lim, R.; Jansson, T.; McIntyre, H.D.; et al. Adipose Tissue Exosomal Proteomic Profile Reveals a Role on Placenta Glucose Metabolism in Gestational Diabetes Mellitus. J. Clin. Endocrinol. Metab. 2019, 104, 1735–1752. [Google Scholar] [CrossRef]
- Baeyens, L.; Hindi, S.; Sorenson, R.L.; German, M.S. beta-Cell adaptation in pregnancy. Diabetes Obes. Metab. 2016, 18 (Suppl. 1), 63–70. [Google Scholar] [CrossRef] [Green Version]
- Prentki, M.; Nolan, C.J. Islet beta cell failure in type 2 diabetes. J. Clin. Investig. 2006, 116, 1802–1812. [Google Scholar] [CrossRef] [Green Version]
- Moyce, B.L.; Dolinsky, V.W. Maternal beta-Cell Adaptations in Pregnancy and Placental Signalling: Implications for Gestational Diabetes. Int. J. Mol. Sci. 2018, 19, 3467. [Google Scholar] [CrossRef] [Green Version]
- Ehses, J.A.; Perren, A.; Eppler, E.; Ribaux, P.; Pospisilik, J.A.; Maor-Cahn, R.; Gueripel, X.; Ellingsgaard, H.; Schneider, M.K.; Biollaz, G.; et al. Increased number of islet-associated macrophages in type 2 diabetes. Diabetes 2007, 56, 2356–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordmann, T.M.; Dror, E.; Schulze, F.; Traub, S.; Berishvili, E.; Barbieux, C.; Boni-Schnetzler, M.; Donath, M.Y. The Role of Inflammation in beta-cell Dedifferentiation. Sci. Rep. 2017, 7, 6285. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Chen, B.; Huang, S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018, 418, 41–50. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soifer, H.S.; Rossi, J.J.; Saetrom, P. MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 2007, 15, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Pilpel, Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006, 7, 1216–1222. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018, 60, 25. [Google Scholar] [CrossRef] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Filonov, G.S.; Noto, J.J.; Schmidt, C.A.; Hatkevich, T.L.; Wen, Y.; Jaffrey, S.R.; Matera, A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015, 21, 1554–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Guarino, E.; Delli Poggi, C.; Grieco, G.E.; Cenci, V.; Ceccarelli, E.; Crisci, I.; Sebastiani, G.; Dotta, F. Circulating MicroRNAs as Biomarkers of Gestational Diabetes Mellitus: Updates and Perspectives. Int. J. Endocrinol. 2018, 2018, 6380463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Song, L.; Zhou, L.; Wu, J.; Sheng, C.; Chen, H.; Liu, Y.; Gao, S.; Huang, W. A MicroRNA Signature in Gestational Diabetes Mellitus Associated with Risk of Macrosomia. Cell Physiol. Biochem 2015, 37, 243–252. [Google Scholar] [CrossRef]
- Faruq, O.; Vecchione, A. microRNA: Diagnostic Perspective. Front. Med. (Lausanne) 2015, 2, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Gao, G.; Cao, Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis. Markers 2016, 2016, 9085195. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, M.; Kolluru, G.K.; Ahmed, A. Small Molecule, Big Prospects: MicroRNA in Pregnancy and Its Complications. J. Pregnancy 2017, 2017, 6972732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayabalan, N.; Nair, S.; Nuzhat, Z.; Rice, G.E.; Zuniga, F.A.; Sobrevia, L.; Leiva, A.; Sanhueza, C.; Gutierrez, J.A.; Lappas, M.; et al. Cross Talk between Adipose Tissue and Placenta in Obese and Gestational Diabetes Mellitus Pregnancies via Exosomes. Front. Endocrinol. (Lausanne) 2017, 8, 239. [Google Scholar] [CrossRef] [Green Version]
- Grygiel-Gorniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications—A review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zhang, T.; Shi, Z.; Ding, H.; Ling, X. MicroRNA-518d regulates PPARalpha protein expression in the placentas of females with gestational diabetes mellitus. Mol. Med. Rep. 2014, 9, 2085–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muralimanoharan, S.; Maloyan, A.; Myatt, L. Mitochondrial function and glucose metabolism in the placenta with gestational diabetes mellitus: Role of miR-143. Clin. Sci. (London) 2016, 130, 931–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, I.J.; Scheele, C.; Keller, P.; Nielsen, A.R.; Remenyi, J.; Fischer, C.P.; Roder, K.; Babraj, J.; Wahlestedt, C.; Hutvagner, G.; et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, H.F.; Jin, X.H.; Cao, Z.F.; Shi, T.; Ma, X. MiR-98 is involved in rat embryo implantation by targeting Bcl-xl. FEBS Lett. 2014, 588, 574–583. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Xie, N.; Li, Y.; Wang, P.; Zhang, C.; Li, Q.; Liu, X.; Deng, J.; Zhang, C.; Lv, C. Upregulation of TRB2 induced by miR-98 in the early lesions of large artery of type-2 diabetic rat. Mol. Cell Biochem 2012, 361, 305–314. [Google Scholar] [CrossRef]
- Cao, J.L.; Zhang, L.; Li, J.; Tian, S.; Lv, X.D.; Wang, X.Q.; Su, X.; Li, Y.; Hu, Y.; Ma, X.; et al. Up-regulation of miR-98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci. Rep. 2016, 6, 32268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.; Jayabalan, N.; Guanzon, D.; Palma, C.; Scholz-Romero, K.; Elfeky, O.; Zuniga, F.; Ormazabal, V.; Diaz, E.; Rice, G.E.; et al. Human placental exosomes in gestational diabetes mellitus carry a specific set of miRNAs associated with skeletal muscle insulin sensitivity. Clin. Sci. (London) 2018, 132, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Ngo, C.; Jayabalan, N.; Salomon, C.; Lappas, M. Molecular pathways disrupted by gestational diabetes mellitus. J. Mol. Endocrinol. 2019, 63, R51–R72. [Google Scholar] [CrossRef] [Green Version]
- Stirm, L.; Huypens, P.; Sass, S.; Batra, R.; Fritsche, L.; Brucker, S.; Abele, H.; Hennige, A.M.; Theis, F.; Beckers, J.; et al. Maternal whole blood cell miRNA-340 is elevated in gestational diabetes and inversely regulated by glucose and insulin. Sci. Rep. 2018, 8, 1366. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Han, A.; Chen, L.; Sun, J.; Lin, Z.; Zhang, X.; Ren, X. Paip1 overexpression is involved in the progression of gastric cancer and predicts shorter survival of diagnosed patients. Onco Targets Ther. 2019, 12, 6565–6576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.; Feng, J.; Cheng, F.; Cui, X.; Gao, L.; Chen, Y.; Wang, F.; Zhong, T.; Li, Y.; Liu, L. Circular RNA expression profiles in placental villi from women with gestational diabetes mellitus. Biochem. Biophys. Res. Commun. 2018, 498, 743–750. [Google Scholar] [CrossRef]
- Boyle, K.E.; Hwang, H.; Janssen, R.C.; DeVente, J.M.; Barbour, L.A.; Hernandez, T.L.; Mandarino, L.J.; Lappas, M.; Friedman, J.E. Gestational diabetes is characterized by reduced mitochondrial protein expression and altered calcium signaling proteins in skeletal muscle. PLoS ONE 2014, 9, e106872. [Google Scholar] [CrossRef] [Green Version]
- Crescioli, C.; Sturli, N.; Sottili, M.; Bonini, P.; Lenzi, A.; Di Luigi, L. Insulin-like effect of the phosphodiesterase type 5 inhibitor tadalafil onto male human skeletal muscle cells. J. Endocrinol. Investig. 2013, 36, 1020–1026. [Google Scholar] [CrossRef]
- Liong, S.; Lappas, M. Activation of AMPK improves inflammation and insulin resistance in adipose tissue and skeletal muscle from pregnant women. J. Physiol. Biochem. 2015, 71, 703–717. [Google Scholar] [CrossRef]
- Fox, C.S.; Massaro, J.M.; Hoffmann, U.; Pou, K.M.; Maurovich-Horvat, P.; Liu, C.Y.; Vasan, R.S.; Murabito, J.M.; Meigs, J.B.; Cupples, L.A.; et al. Abdominal visceral and subcutaneous adipose tissue compartments: Association with metabolic risk factors in the Framingham Heart Study. Circulation 2007, 116, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Akbay, E.; Tiras, M.B.; Yetkin, I.; Toruner, F.; Ersoy, R.; Uysal, S.; Ayvaz, G. Insulin secretion and insulin sensitivity in normal pregnancy and gestational diabetes mellitus. Gynecol. Endocrinol. 2003, 17, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Ropero, A.B.; Alonso-Magdalena, P.; Quesada, I.; Nadal, A. The role of estrogen receptors in the control of energy and glucose homeostasis. Steroids 2008, 73, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Nadal, A.; Alonso-Magdalena, P.; Soriano, S.; Quesada, I.; Ropero, A.B. The pancreatic beta-cell as a target of estrogens and xenoestrogens: Implications for blood glucose homeostasis and diabetes. Mol. Cell Endocrinol. 2009, 304, 63–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso-Magdalena, P.; Ropero, A.B.; Carrera, M.P.; Cederroth, C.R.; Baquie, M.; Gauthier, B.R.; Nef, S.; Stefani, E.; Nadal, A. Pancreatic insulin content regulation by the estrogen receptor ER alpha. PLoS ONE 2008, 3, e2069. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Li, P.; Li, L. Whole transcriptome expression profiles in placenta samples from women with gestational diabetes mellitus. J. Diabetes Investig. 2020. [Google Scholar] [CrossRef] [Green Version]
- Bellini, L.; Campana, M.; Mahfouz, R.; Carlier, A.; Veret, J.; Magnan, C.; Hajduch, E.; Le Stunff, H. Targeting sphingolipid metabolism in the treatment of obesity/type 2 diabetes. Expert Opin. Ther. Targets 2015, 19, 1037–1050. [Google Scholar] [CrossRef]
- Batista, T.M.; Garcia-Martin, R.; Cai, W.; Konishi, M.; O’Neill, B.T.; Sakaguchi, M.; Kim, J.H.; Jung, D.Y.; Kim, J.K.; Kahn, C.R. Multi-dimensional Transcriptional Remodeling by Physiological Insulin In Vivo. Cell Rep. 2019, 26, 3429–3443.e3. [Google Scholar] [CrossRef] [Green Version]
- Moon, M.K.; Kang, G.H.; Kim, H.H.; Han, S.K.; Koo, Y.D.; Cho, S.W.; Kim, Y.A.; Oh, B.C.; Park do, J.; Chung, S.S.; et al. Thyroid-stimulating hormone improves insulin sensitivity in skeletal muscle cells via cAMP/PKA/CREB pathway-dependent upregulation of insulin receptor substrate-1 expression. Mol. Cell Endocrinol. 2016, 436, 50–58. [Google Scholar] [CrossRef]
- Banerjee, R.R.; Cyphert, H.A.; Walker, E.M.; Chakravarthy, H.; Peiris, H.; Gu, X.; Liu, Y.; Conrad, E.; Goodrich, L.; Stein, R.W.; et al. Gestational Diabetes Mellitus From Inactivation of Prolactin Receptor and MafB in Islet beta-Cells. Diabetes 2016, 65, 2331–2341. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wang, S.; Li, H.; Wan, J.; Zhou, Q.; Zhou, Y.; Zhang, C. microRNA-96 protects pancreatic beta-cell function by targeting PAK1 in gestational diabetes mellitus. Biofactors 2018, 44, 539–547. [Google Scholar] [CrossRef]
- Ahn, M.; Yoder, S.M.; Wang, Z.; Oh, E.; Ramalingam, L.; Tunduguru, R.; Thurmond, D.C. The p21-activated kinase (PAK1) is involved in diet-induced beta cell mass expansion and survival in mice and human islets. Diabetologia 2016, 59, 2145–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebastiani, G.; Guarino, E.; Grieco, G.E.; Formichi, C.; Delli Poggi, C.; Ceccarelli, E.; Dotta, F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. (Lausanne) 2017, 8, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annicotte, J.S.; Blanchet, E.; Chavey, C.; Iankova, I.; Costes, S.; Assou, S.; Teyssier, J.; Dalle, S.; Sardet, C.; Fajas, L. The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat. Cell Biol. 2009, 11, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Oh, E.; Thurmond, D.C. Glucose-stimulated Cdc42 signaling is essential for the second phase of insulin secretion. J. Biol. Chem. 2007, 282, 9536–9546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, Y.; Guo, N.; He, T.; Ji, J.; Li, Z.; Huang, P. miR-410 enhanced hESC-derived pancreatic endoderm transplant to alleviate gestational diabetes mellitus. J. Mol. Endocrinol. 2015, 55, 219–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ainscow, E.K.; Zhao, C.; Rutter, G.A. Acute overexpression of lactate dehydrogenase-A perturbs beta-cell mitochondrial metabolism and insulin secretion. Diabetes 2000, 49, 1149–1155. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef]
- Cao, Y.L.; Jia, Y.J.; Xing, B.H.; Shi, D.D.; Dong, X.J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J. Obstet. Gynaecol. Res. 2017, 43, 974–981. [Google Scholar] [CrossRef]
- Wander, P.L.; Boyko, E.J.; Hevner, K.; Parikh, V.J.; Tadesse, M.G.; Sorensen, T.K.; Williams, M.A.; Enquobahrie, D.A. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res. Clin. Pract. 2017, 132, 1–9. [Google Scholar] [CrossRef]
- Hocaoglu, M.; Demirer, S.; Senturk, H.; Turgut, A.; Komurcu-Bayrak, E. Differential expression of candidate circulating microRNAs in maternal blood leukocytes of the patients with preeclampsia and gestational diabetes mellitus. Pregnancy Hypertens. 2019, 17, 5–11. [Google Scholar] [CrossRef]
- Tagoma, A.; Alnek, K.; Kirss, A.; Uibo, R.; Haller-Kikkatalo, K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018, 672, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Jump, D.B. Fatty acid regulation of hepatic lipid metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 115–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014, 588, 3939–3946. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Dong, J.; Jiang, T.; Shi, Z.; Yu, B.; Zhu, Y.; Chen, D.; Xu, J.; Huo, R.; Dai, J.; et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE 2011, 6, e23925. [Google Scholar] [CrossRef]
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352. [Google Scholar] [CrossRef]
- Yoffe, L.; Polsky, A.; Gilam, A.; Raff, C.; Mecacci, F.; Ognibene, A.; Crispi, F.; Gratacos, E.; Kanety, H.; Mazaki-Tovi, S.; et al. Early diagnosis of gestational diabetes mellitus using circulating microRNAs. Eur. J. Endocrinol. 2019, 181, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Lamadrid-Romero, M.; Solis, K.H.; Cruz-Resendiz, M.S.; Perez, J.E.; Diaz, N.F.; Flores-Herrera, H.; Garcia-Lopez, G.; Perichart, O.; Reyes-Munoz, E.; Arenas-Huertero, F.; et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci. Res. 2018, 130, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wu, H.; Wang, F.; Ye, M.; Zhu, H.; Bu, S. Long non-coding RNA MALAT1 expression in patients with gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2018, 140, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, X.; Li, C.; Zhang, W.; Lv, Y.; Wang, L.; Wu, L.; Meng, L.; Fan, Y.; Ding, H.; et al. Down-regulated long non-coding RNA PVT1 contributes to gestational diabetes mellitus and preeclampsia via regulation of human trophoblast cells. Biomed. Pharmacother. 2019, 120, 109501. [Google Scholar] [CrossRef]
- Ye, H.H.; Yang, S.H.; Zhang, Y. MEG3 damages fetal endothelial function induced by gestational diabetes mellitus via AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8553–8560. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, C.; Long, W.; Ding, H.; Shen, R. Microarray Expression Profile Analysis of Long Non-Coding RNAs in Umbilical Cord Plasma Reveals their Potential Role in Gestational Diabetes-Induced Macrosomia. Cell Physiol. Biochem. 2015, 36, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wu, J.; Zhao, Z.; Wang, J.; Chen, Z. Circulating LncRNA Serve as Fingerprint for Gestational Diabetes Mellitus Associated with Risk of Macrosomia. Cell Physiol. Biochem. 2018, 48, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Guo, F.; Zhang, Y.; Liu, X.M.; Xiang, Y.Q.; Zhang, C.; Liu, Z.W.; Sheng, J.Z.; Huang, H.F.; Zhang, J.Y.; et al. Integrated Transcriptome Sequencing Analysis Reveals Role of miR-138-5p/ TBL1X in Placenta from Gestational Diabetes Mellitus. Cell Physiol. Biochem. 2018, 51, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Liu, F.Q.; Yan, R.; Yi, H.M.; Zhang, T.; Wang, G.Y.; Li, Y.; Jiang, N. Downregulation of miR-610 promotes proliferation and tumorigenicity and activates Wnt/beta-catenin signaling in human hepatocellular carcinoma. Mol. Cancer 2014, 13, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knofler, M.; Pollheimer, J. Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.Y.; Li, M.Q.; Li, H.P. High glucose suppresses the viability and proliferation of HTR8/SVneo cells through regulation of the miR137/PRKAA1/IL6 axis. Int. J. Mol. Med. 2018, 42, 799–810. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.Y.; Li, H.P.; Li, M.Q. High glucose induces dysfunction of human umbilical vein endothelial cells by upregulating miR-137 in gestational diabetes mellitus. Microvasc. Res. 2018, 118, 90–100. [Google Scholar] [CrossRef]
- Smits, M.; Mir, S.E.; Nilsson, R.J.; van der Stoop, P.M.; Niers, J.M.; Marquez, V.E.; Cloos, J.; Breakefield, X.O.; Krichevsky, A.M.; Noske, D.P.; et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PLoS ONE 2011, 6, e16282. [Google Scholar] [CrossRef]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [Green Version]
- Mitic, T.; Caporali, A.; Floris, I.; Meloni, M.; Marchetti, M.; Urrutia, R.; Angelini, G.D.; Emanueli, C. EZH2 modulates angiogenesis in vitro and in a mouse model of limb ischemia. Mol. Ther. 2015, 23, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Han, H.D.; Mangala, L.S.; Ali-Fehmi, R.; Newton, C.S.; Ozbun, L.; Armaiz-Pena, G.N.; Hu, W.; Stone, R.L.; Munkarah, A.; et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell 2010, 18, 185–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kottakis, F.; Polytarchou, C.; Foltopoulou, P.; Sanidas, I.; Kampranis, S.C.; Tsichlis, P.N. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol. Cell 2011, 43, 285–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreger, H.; Ludwig, A.; Weller, A.; Stangl, V.; Baumann, G.; Meiners, S.; Stangl, K. Epigenetic regulation of cell adhesion and communication by enhancer of zeste homolog 2 in human endothelial cells. Hypertension 2012, 60, 1176–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floris, I.; Descamps, B.; Vardeu, A.; Mitic, T.; Posadino, A.M.; Shantikumar, S.; Sala-Newby, G.; Capobianco, G.; Mangialardi, G.; Howard, L.; et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 664–674. [Google Scholar] [CrossRef] [Green Version]
- Favaro, E.; Miceli, I.; Bussolati, B.; Schmitt-Ney, M.; Cavallo Perin, P.; Camussi, G.; Zanone, M.M. Hyperglycemia induces apoptosis of human pancreatic islet endothelial cells: Effects of pravastatin on the Akt survival pathway. Am. J. Pathol. 2008, 173, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Forbes, K.; Westwood, M. Maternal growth factor regulation of human placental development and fetal growth. J. Endocrinol. 2010, 207, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Scifres, C.M.; Nelson, D.M. Intrauterine growth restriction, human placental development and trophoblast cell death. J. Physiol. 2009, 587, 3453–3458. [Google Scholar] [CrossRef] [PubMed]
- Er, E.E.; Mendoza, M.C.; Mackey, A.M.; Rameh, L.E.; Blenis, J. AKT facilitates EGFR trafficking and degradation by phosphorylating and activating PIKfyve. Sci. Signal 2013, 6, ra45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Bezprozvannaya, S.; Shelton, J.M.; Frisard, M.I.; Hulver, M.W.; McMillan, R.P.; Wu, Y.; Voelker, K.A.; Grange, R.W.; Richardson, J.A.; et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J. Clin. Investig. 2011, 121, 3258–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Williams, A.H.; Kim, Y.; McAnally, J.; Bezprozvannaya, S.; Sutherland, L.B.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. USA 2007, 104, 20844–20849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torella, D.; Iaconetti, C.; Catalucci, D.; Ellison, G.M.; Leone, A.; Waring, C.D.; Bochicchio, A.; Vicinanza, C.; Aquila, I.; Curcio, A.; et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ. Res. 2011, 109, 880–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mughal, W.; Nguyen, L.; Pustylnik, S.; da Silva Rosa, S.C.; Piotrowski, S.; Chapman, D.; Du, M.; Alli, N.S.; Grigull, J.; Halayko, A.J.; et al. A conserved MADS-box phosphorylation motif regulates differentiation and mitochondrial function in skeletal, cardiac, and smooth muscle cells. Cell Death Dis. 2015, 6, e1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Care, A.; Catalucci, D.; Felicetti, F.; Bonci, D.; Addario, A.; Gallo, P.; Bang, M.L.; Segnalini, P.; Gu, Y.; Dalton, N.D.; et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007, 13, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Filardi, T.; Ghinassi, B.; Di Baldassarre, A.; Tanzilli, G.; Morano, S.; Lenzi, A.; Basili, S.; Crescioli, C. Cardiomyopathy Associated with Diabetes: The Central Role of the Cardiomyocyte. Int. J. Mol. Sci. 2019, 20, 3299. [Google Scholar] [CrossRef] [Green Version]
- Yussman, M.G.; Toyokawa, T.; Odley, A.; Lynch, R.A.; Wu, G.; Colbert, M.C.; Aronow, B.J.; Lorenz, J.N.; Dorn, G.W., 2nd. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat. Med. 2002, 8, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Lehnen, H.; Zechner, U.; Haaf, T. Epigenetics of gestational diabetes mellitus and offspring health: The time for action is in early stages of life. Mol. Hum. Reprod. 2013, 19, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnevale, R.; Raparelli, V.; Nocella, C.; Bartimoccia, S.; Novo, M.; Severino, A.; De Falco, E.; Cammisotto, V.; Pasquale, C.; Crescioli, C.; et al. Gut-derived endotoxin stimulates factor VIII secretion from endothelial cells. Implications for hypercoagulability in cirrhosis. J. Hepatol. 2017, 67, 950–956. [Google Scholar] [CrossRef]
- Giannattasio, S.; Corinaldesi, C.; Colletti, M.; Di Luigi, L.; Antinozzi, C.; Filardi, T.; Scolletta, S.; Basili, S.; Lenzi, A.; Morano, S.; et al. The phosphodiesterase 5 inhibitor sildenafil decreases the proinflammatory chemokine IL-8 in diabetic cardiomyopathy: In vivo and in vitro evidence. J. Endocrinol. Investig. 2019, 42, 715–725. [Google Scholar] [CrossRef] [Green Version]
- Mrizak, I.; Arfa, A.; Fekih, M.; Debbabi, H.; Bouslema, A.; Boumaiza, I.; Zaouali, M.; Khan, N.A.; Tabka, Z. Inflammation and impaired endothelium-dependant vasodilatation in non obese women with gestational diabetes mellitus: Preliminary results. Lipids Health Dis. 2013, 12, 93. [Google Scholar] [CrossRef] [Green Version]
- Strutz, J.; Cvitic, S.; Hackl, H.; Kashofer, K.; Appel, H.M.; Thuringer, A.; Desoye, G.; Koolwijk, P.; Hiden, U. Gestational diabetes alters microRNA signatures in human feto-placental endothelial cells depending on fetal sex. Clin. Sci. (London) 2018, 132, 2437–2449. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Yao, H.; Hu, G.; Chen, X.; Lund, A.K.; Buch, S. HIV Tat induces expression of ICAM-1 in HUVECs: Implications for miR-221/-222 in HIV-associated cardiomyopathy. PLoS ONE 2013, 8, e60170. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Gong, A.Y.; Liu, J.; Zhou, R.; Deng, C.; Chen, X.M. miR-221 suppresses ICAM-1 translation and regulates interferon-gamma-induced ICAM-1 expression in human cholangiocytes. Am. J. Physiol. Gastrointest Liver Physiol. 2010, 298, G542–G550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Perez, F.I.; Hiden, U.; Gauster, M.; Lang, I.; Konya, V.; Heinemann, A.; Logl, J.; Saffery, R.; Desoye, G.; Cvitic, S. Post-transcriptional down regulation of ICAM-1 in feto-placental endothelium in GDM. Cell Adh. Migr. 2016, 10, 18–27. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Vishwanath, A.; Jiang, S.; Mallappa, A.; Teague, A.M.; Takahashi, Y.; Thompson, D.M.; Chernausek, S.D. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin. Sci. (London) 2016, 130, 1955–1967. [Google Scholar] [CrossRef] [Green Version]
- Houshmand-Oeregaard, A.; Schrolkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.C.B.; Broholm, C.; Mathiesen, E.R.; Clausen, T.D.; Vaag, A.; et al. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771. [Google Scholar] [CrossRef]
- Bork-Jensen, J.; Scheele, C.; Christophersen, D.V.; Nilsson, E.; Friedrichsen, M.; Fernandez-Twinn, D.S.; Grunnet, L.G.; Litman, T.; Holmstrom, K.; Vind, B.; et al. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: Results from studies of twins with and without type 2 diabetes. Diabetologia 2015, 58, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Terauchi, Y.; Tsuji, Y.; Satoh, S.; Minoura, H.; Murakami, K.; Okuno, A.; Inukai, K.; Asano, T.; Kaburagi, Y.; Ueki, K.; et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat. Genet. 1999, 21, 230–235. [Google Scholar] [CrossRef]
- Sun, L.L.; Jiang, B.G.; Li, W.T.; Zou, J.J.; Shi, Y.Q.; Liu, Z.M. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res. Clin. Pract. 2011, 91, 94–100. [Google Scholar] [CrossRef]
- Marcondes, J.P.C.; Andrade, P.F.B.; Savio, A.L.V.; Silveira, M.A.D.; Rudge, M.V.C.; Salvadori, D.M.F. BCL2 and miR-181a transcriptional alterations in umbilical-cord blood cells can be putative biomarkers for obesity. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 836, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Rippo, M.R.; Olivieri, F.; Monsurro, V.; Prattichizzo, F.; Albertini, M.C.; Procopio, A.D. MitomiRs in human inflamm-aging: A hypothesis involving miR-181a, miR-34a and miR-146a. Exp. Gerontol. 2014, 56, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Fornes, D.; White, V.; Higa, R.; Heinecke, F.; Capobianco, E.; Jawerbaum, A. Sex-dependent changes in lipid metabolism, PPAR pathways and microRNAs that target PPARs in the fetal liver of rats with gestational diabetes. Mol. Cell Endocrinol. 2018, 461, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
- Laprovitera, N.; Grzes, M.; Porcellini, E.; Ferracin, M. Cancer Site-Specific Multiple microRNA Quantification by Droplet Digital PCR. Front. Oncol. 2018, 8, 447. [Google Scholar] [CrossRef] [PubMed]
| Study | Groups | Stage of Pregnancy | Source | lncRNA | Target miRNA | Target Pathway | Role/Biological Function |
|---|---|---|---|---|---|---|---|
| Zhang et al. [109] | 50 GDM 47 CTRL | 36th–40th week | Serum | MALAT1 (↑) | - | - | Circulating biomarker of GDM |
| Wang et al. [110] | 13 GDM 15 PE 13 CTRL | Term | Placenta | PVT1 (↓) | miR-10-5p miR-423-5p miR-374b-5p miR-378b miR-150-5p miR-194-5p miR-3184 | PI3K/AKT | Trophoblast dysfunction (↓ proliferation and migration) |
| Ye et al. [111] | 16 GDM 18 CTRL | Term | HUVECs | MEG3 (↑) | miR-370-3p | PI3K/AKT | Endothelial dysfunction (↓ tube formation and migration) |
| Shi et al. [112] | 30 GDM-M30 CTRL-M | Term | Plasma (HUVECs) | ENST00000552367 (↑) | - | TADG51/IGF1 | ↓ Apoptosis |
| Lu et al. [113] | 150 GDM-M 150 CTRL-M 150 CTRL 150 Np | Early pregnancy | Plasma | XLOC_014172 (↑) RP11-230G5.2 (↑) | - | - | Circulating biomarkers of macrosomia in GDM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filardi, T.; Catanzaro, G.; Mardente, S.; Zicari, A.; Santangelo, C.; Lenzi, A.; Morano, S.; Ferretti, E. Non-Coding RNA: Role in Gestational Diabetes Pathophysiology and Complications. Int. J. Mol. Sci. 2020, 21, 4020. https://doi.org/10.3390/ijms21114020
Filardi T, Catanzaro G, Mardente S, Zicari A, Santangelo C, Lenzi A, Morano S, Ferretti E. Non-Coding RNA: Role in Gestational Diabetes Pathophysiology and Complications. International Journal of Molecular Sciences. 2020; 21(11):4020. https://doi.org/10.3390/ijms21114020
Chicago/Turabian StyleFilardi, Tiziana, Giuseppina Catanzaro, Stefania Mardente, Alessandra Zicari, Carmela Santangelo, Andrea Lenzi, Susanna Morano, and Elisabetta Ferretti. 2020. "Non-Coding RNA: Role in Gestational Diabetes Pathophysiology and Complications" International Journal of Molecular Sciences 21, no. 11: 4020. https://doi.org/10.3390/ijms21114020