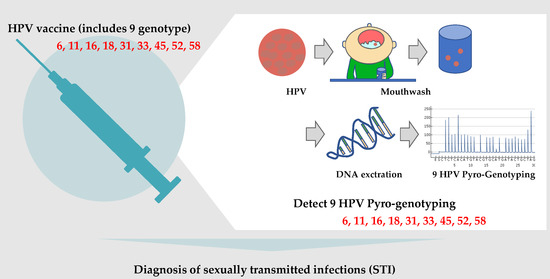
Mouthwash-Based Highly Sensitive Pyro-Genotyping for Nine Sexually Transmitted Human Papilloma Virus Genotypes
Abstract
:1. Introduction
2. Results
2.1. Volume and Quality of Genomic DNA Recovered from Mouthwash
2.2. Amplification and Direct Sequencing of the 18 HPV Genotype-Harboring Plasmids from HeLa Cells
2.3. Amplification of Genes from HPV in the Clinical Mouthwash Waste Samples
2.4. Pyrosequencing Was Suitable for Genotyping Nine HPV Plasmids from HeLa Cells and Clinical Mouthwash Samples
2.5. Pyrosequencing Approach Was More Sensitive than the Conventional HPV-DNA Method
3. Discussion
4. Materials and Methods
4.1. Characteristics of the Patient Samples
4.2. Collection of Mouthwash
4.3. Construction of Plasmid DNAs Containing the Variable Sequence of the 18 HPV Genotypes
4.4. Direct Sequencing Using Plasmid DNAs Harboring the 18 HPV Genotypes from HeLa Cells
4.5. Quantitative Pyrosequencing Analysis Using Plasmid DNAs Harboring the 18 HPV Genotypes from HeLa Cells and Clinical Mouthwash Samples
4.6. HPV Detection using the Conventional HPV-DNA Method and Clinical Mouthwash Samples
4.7. HPV Detection using the Conventional HPV Genotyping Method and Clinical Mouthwash Samples
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| STI | Sexually transmitted infections |
| HPV | Human papilloma virus |
| Pyro-genotyping | Pyrosequencing-based genotyping |
| HC2 | Hybrid capture 2 |
References
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. USA 1983, 80, 3812–3815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagami, W.; Nagase, S.; Takahashi, F.; Ino, K.; Hachisuga, T.; Aoki, D.; Katabuchi, H. Clinical statistics of gynecologic cancers in Japan. J. Gynecol. Oncol. 2017, 28, e32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.S.; Lindsay, L.; Hoots, B.; Keys, J.; Franceschi, S.; Winer, R.; Clifford, G.M. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: A meta-analysis update. Int. J. Cancer 2007, 121, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, C.M.; Hunt, W.C.; Schiffman, M.; Castle, P.E. Human papillomavirus genotypes and the cumulative 2-year risk of cervical precancer. J. Infect. Dis. 2006, 194, 1291–1299. [Google Scholar] [CrossRef] [Green Version]
- Fortes, H.R.; von Ranke, F.M.; Escuissato, D.L.; Araujo Neto, C.A.; Zanetti, G.; Hochhegger, B.; Souza, C.A.; Marchiori, E. Recurrent respiratory papillomatosis: A state-of-the-art review. Respir. Med. 2017, 126, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Parisi, S.G.; Basso, M.; Scaggiante, R.; Andreis, S.; Mengoli, C.; Cruciani, M.; Del, V.C.; Menegotto, N.; Zago, D.; Sarmati, L.; et al. Oral and anal high-risk human papilloma virus infection in HIV-positive men who have sex with men over a 24-month longitudinal study: Complexity and vaccine implications. BMC Public Health 2019, 19, 645. [Google Scholar] [CrossRef] [PubMed]
- Van Bilsen, W.P.H.; Kovaleva, A.; Bleeker, M.C.G.; King, A.J.; Bruisten, S.M.; Brokking, W.; De Vries, H.J.C.; Meijer, C.J.L.M.; Loeff, M.F.S.V.D. HPV infections and flat penile lesions of the penis in men who have sex with men. Papillomavirus Res. 2019, 8, 100173. [Google Scholar] [CrossRef]
- Majewski, S.; Jablonska, S. Possible involvement of epidermodysplasia verruciformis human papillomaviruses in the immunopathogenesis of psoriasis: A proposed hypothesis. Exp. Dermatol. 2003, 12, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Paz, I.B.; Cook, N.; Odom-Maryon, T.; Xie, Y.; Wilczynski, S.P. Human papillomavirus (HPV) in head and neck cancer. An association of HPV 16 with squamous cell carcinoma of Waldeyer’s tonsillar ring. Cancer 1997, 79, 595–604. [Google Scholar] [CrossRef]
- Harper, D.M.; Franco, E.L.; Wheeler, C.M.; Moscicki, A.B.; Romanowski, B.; Roteli-Martins, C.M.; Jenkins, D.; Schuind, A.; Costa Clemens, S.A.; Dubin, G. Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: Follow-up from a randomised control trial. Lancet 2006, 367, 1247–1255. [Google Scholar] [CrossRef] [Green Version]
- Joura, E.A.; Leodolter, S.; Hernandez-Avila, M.; Wheeler, C.M.; Perez, G.; Koutsky, L.A.; Garland, S.M.; Harper, D.M.; Tang, G.W.; Ferris, D.G.; et al. Efficacy of a quadrivalent prophylactic human papillomavirus (types 6, 11, 16, and 18) L1 virus-like-particle vaccine against high-grade vulval and vaginal lesions: A combined analysis of three randomised clinical trials. Lancet 2007, 369, 1693–1702. [Google Scholar] [CrossRef]
- Olsson, S.E.; Villa, L.L.; Costa, R.L.; Petta, C.A.; Andrade, R.P.; Malm, C.; Iversen, O.E.; Hoye, J.; Steinwall, M.; Riis-Johannessen, G.; et al. Induction of immune memory following administration of a prophylactic quadrivalent human papillomavirus (HPV) types 6/11/16/18 L1 virus-like particle (VLP) vaccine. Vaccine 2007, 25, 4931–4939. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Joura, E.A.; Giuliano, A.R.; Iversen, O.E.; de Andrade, R.P.; Ault, K.A.; Bartholomew, D.; Cestero, R.M.; Fedrizzi, E.N.; Hirschberg, A.L.; et al. Final efficacy, immunogenicity, and safety analyses of a nine-valent human papillomavirus vaccine in women aged 16-26 years: A randomised, double-blind trial. Lancet 2017, 390, 2143–2159. [Google Scholar] [CrossRef]
- Stanley, M.; Lowy, D.R.; Frazer, I. Chapter 12: Prophylactic HPV vaccines: Underlying mechanisms. Vaccine 2006, 24, S106–S113. [Google Scholar] [CrossRef]
- Taniguchi, M.; Ueda, Y.; Yagi, A.; Ikeda, S.; Endo, M.; Tomimatsu, T.; Nakayama, T.; Sekine, M.; Enomoto, T.; Kimura, T. Cervical cancer screening rate differs by HPV vaccination status: An interim analysis. Vaccine 2019, 37, 4424–4426. [Google Scholar] [CrossRef]
- Inoue, M.; Nakazawa, A.; Fujita, M.; Tanizawa, O. Human papillomavirus (HPV) type 16 in semen of partners of women with HPV infection. Lancet 1992, 339, 1114–1115. [Google Scholar] [CrossRef]
- Halkitis, P.N.; Valera, P.; LoSchiavo, C.E.; Goldstone, S.E.; Kanztanou, M.; Maiolatesi, A.J.; Ompad, D.C.; Greene, R.E.; Kapadia, F. Human papillomavirus vaccination and infection in young sexual minority men: The P18 cohort study. AIDS Patient Care STDS. 2019, 33, 149–156. [Google Scholar] [CrossRef]
- Woodman, C.B.J.; Collins, S.I.; Young, L.S. The natural history of cervical HPV infection: Unresolved issues. Nat. Rev. Cancer 2007, 7, 11–22. [Google Scholar] [CrossRef]
- Patterson, N.A.; Smith, J.L.; Ozbun, M.A. Human papillomavirus type 31b infection of human keratinocytes does not require heparan sulfate. J. Virol. 2005, 79, 6838–6847. [Google Scholar] [CrossRef] [Green Version]
- Zur Hausen, H. Papillomavirus infections—A major cause of human cancers. Biochim. Biophys. Acta Rev. Cancer 1996, 1288, F55–F78. [Google Scholar] [CrossRef]
- Antonsson, A.; Cornford, M.; Perry, S.; Davis, M.; Dunne, M.P.; Whiteman, D.C. Prevalence and risk factors for oral HPV infection in young Australians. PLoS ONE. 2014, 9, e91761. [Google Scholar] [CrossRef] [PubMed]
- Syrjanen, S. Oral manifestations of human papillomavirus infections. Eur. J. Oral Sci. 2018, 126, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Nobbenhuis, M.A.; Walboomers, J.M.; Helmerhorst, T.J.; Rozendaal, L.; Remmink, A.J.; Risse, E.K.; van der Linden, H.C.; Voorhorst, F.J.; Kenemans, P.; Meijer, C.J. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: A prospective study. Lancet 1999, 354, 20–25. [Google Scholar] [CrossRef]
- Clavel, C.; Masure, M.; Bory, J.P.; Putaud, I.; Mangeonjean, C.; Lorenzato, M.; Gabriel, R.; Quereux, C.; Birembaut, P. Hybrid capture II-based human papillomavirus detection, a sensitive test to detect in routine high-grade cervical lesions: A preliminary study on 1518 women. Br. J. Cancer 1999, 80, 1306–1311. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, M.; Huang, B.; Katabi, N.; Migliacci, J.; Bryant, R.; Kaplan, S.; Blackwell, T.; Patel, S.; Yang, L.; Pei, Z.; et al. Detection of HPV related oropharyngeal cancer in oral rinse specimens. Oncotarget 2017, 8, 109393–109401. [Google Scholar] [CrossRef] [Green Version]
- Fakhry, C.; Qeadan, F.; Gilman, R.H.; Yori, P.; Kosek, M.; Patterson, N.; Eisele, D.W.; Gourin, C.G.; Chitguppi, C.; Marks, M.; et al. Oral sampling methods are associated with differences in immune marker concentrations. Laryngoscope 2018, 128, E214–E221. [Google Scholar] [CrossRef]
- Miyamoto, S.; Watanabe, Y.; Oikawa, R.; Ono, S.; Mabe, K.; Kudo, T.; Yamamoto, H.; Itoh, F.; Kato, M.; Sakamoto, N. Analysis of Helicobacter pylori genotypes in clinical gastric wash samples. Tumor Biol. 2016, 37, 10123–10132. [Google Scholar] [CrossRef]
- Baba, S.; Oishi, Y.; Watanabe, Y.; Oikawa, R.; Morita, R.; Yoshida, Y.; Hiraishi, T.; Maehata, T.; Nagase, Y.; Fukuda, Y.; et al. Gastric wash-based molecular testing for antibiotic resistance in Helicobacter pylori. Digestion 2011, 84, 299–305. [Google Scholar] [CrossRef]
- Oikawa, R.; Watanabe, Y.; Miyamoto, S.; Sato, Y.; Ono, S.; Mabe, K.; Yamamoto, H.; Kato, M.; Itoh, F. Enrichment of Helicobacter pylori mutant strains after eradication therapy analyzed by gastric wash–based quantitative pyrosequencing. Tumor Biol. 2017, 39, 1010428317734865. [Google Scholar] [CrossRef] [Green Version]
- Nobre, R.J.; de Almeida, L.P.; Martins, T.C. Complete genotyping of mucosal human papillomavirus using a restriction fragment length polymorphism analysis and an original typing algorithm. J. Clin. Virol. 2008, 42, 13–21. [Google Scholar] [CrossRef] [Green Version]



| Unit No. | DNA Conc. (ng) | Pyrosequencing | Restriction Enzyme-Based Genotyping | ||
|---|---|---|---|---|---|
| Amplification | HPV Genotype | Hybrid Capture | |||
| 1 | 811.9 | ||||
| 2 | 146.1 | Amplified | 18 | 18 | |
| 3 | 156.7 | ||||
| 4 | 708.7 | ||||
| 5 | 67.8 | ||||
| 6 | 117.2 | Amplified | 18 | Unidentified | |
| 7 | 275.5 | ||||
| 8 | 153.1 | ||||
| 9 | 456.2 | ||||
| 10 | 612.4 | ||||
| 11 | 104.5 | ||||
| 12 | 586.7 | ||||
| 13 | 367.6 | Amplified | 16 | Detected | 16 |
| 14 | 128.8 | Amplified | 18 | Unidentified | |
| 15 | 385.1 | ||||
| 16 | 327.5 | ||||
| 17 | 189.9 | ||||
| 18 | 252.4 | ||||
| 19 | 876.1 | ||||
| 20 | 8.4 | ||||
| 21 | 290.9 | ||||
| 22 | 37.4 | ||||
| 23 | 344.7 | Amplified | 11 | Unidentified | |
| 24 | 26.1 | ||||
| 25 | 344.8 | ||||
| 26 | 3.8 | ||||
| 27 | 0.9 | ||||
| 28 | 197.3 | ||||
| 29 | 431.3 | ||||
| 30 | 301.1 | ||||
| 31 | 280.6 | Amplified | 6 | Unidentified | |
| 32 | 275.2 | ||||
| 33 | 486.1 | ||||
| 34 | 380.2 | Amplified | 18 | Unidentified | |
| 35 | 298.4 | Amplified | 18 | Unidentified | |
| 36 | 260.3 | ||||
| 37 | 280.8 | ||||
| 38 | 180.2 | ||||
| 39 | 302.1 | ||||
| 40 | 197.6 | ||||
| 41 | 245.2 | ||||
| 42 | 266.1 | ||||
| 43 | 407.8 | ||||
| 44 | 28.1 | ||||
| 45 | 180.3 | ||||
| 46 | 199.1 | ||||
| 47 | 240.6 | Amplified | 18 | Unidentified | |
| 48 | 150.2 | Amplified | 18 | Unidentified | |
| 49 | 118.2 | ||||
| 50 | 260.8 | Amplified | 18 | Unidentified | |
| 51 | 110.8 | ||||
| 52 | 320.1 | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, Y.; Seto, Y.; Oikawa, R.; Nakazawa, T.; Furuya, H.; Matsui, H.; Hosono, S.; Noike, M.; Inoue, A.; Yamamoto, H.; et al. Mouthwash-Based Highly Sensitive Pyro-Genotyping for Nine Sexually Transmitted Human Papilloma Virus Genotypes. Int. J. Mol. Sci. 2020, 21, 3697. https://doi.org/10.3390/ijms21103697
Watanabe Y, Seto Y, Oikawa R, Nakazawa T, Furuya H, Matsui H, Hosono S, Noike M, Inoue A, Yamamoto H, et al. Mouthwash-Based Highly Sensitive Pyro-Genotyping for Nine Sexually Transmitted Human Papilloma Virus Genotypes. International Journal of Molecular Sciences. 2020; 21(10):3697. https://doi.org/10.3390/ijms21103697
Chicago/Turabian StyleWatanabe, Yoshiyuki, Yukiko Seto, Ritsuko Oikawa, Takara Nakazawa, Hanae Furuya, Hidehito Matsui, Sachiko Hosono, Mika Noike, Akiko Inoue, Hiroyuki Yamamoto, and et al. 2020. "Mouthwash-Based Highly Sensitive Pyro-Genotyping for Nine Sexually Transmitted Human Papilloma Virus Genotypes" International Journal of Molecular Sciences 21, no. 10: 3697. https://doi.org/10.3390/ijms21103697