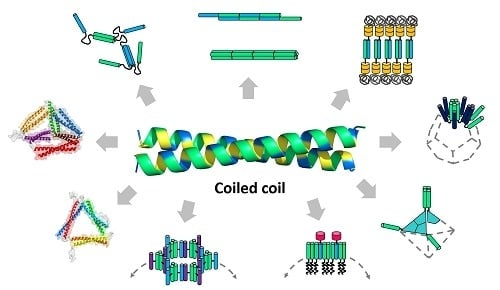
Coiled-Coils: The Molecular Zippers that Self-Assemble Protein Nanostructures
Abstract
:1. Introduction
2. Modular Design Principles
3. Supramolecular Nanostructures Built from Coiled-Coils
3.1. Unbounded Nanostructures
3.2. Discrete Nanoparticles
3.3. Origami Nanostructures
4. Conclusions and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Lupas, A.N.; Bassler, J. Coiled coils–a model system for the 21st century. Trends Biochem. Sci. 2017, 42, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Hai, T.; Curran, T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA 1991, 88, 3720–3724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitby, F.G.; Phillips, G.N., Jr. Crystal structure of tropomyosin at 7 Ångstroms resolution. Proteins Struct. Funct. Bioinform. 2000, 38, 49–59. [Google Scholar] [CrossRef]
- Koubassova, N.; Bershitsky, S.; Tsaturyan, A. Effects of an Interchain Disulfide Bond on Tropomyosin Structure: A Molecular Dynamics Study. Int. J. Mol. Sci. 2018, 19, 3376. [Google Scholar] [CrossRef] [Green Version]
- Mason, J.M.; Arndt, K.M. Coiled coil domains: Stability, specificity, and biological implications. Chembiochem 2004, 5, 170–176. [Google Scholar] [CrossRef]
- Lupas, A.N.; Gruber, M. The structure of α-helical coiled coils. Adv. Protein Chem. 2005, 70, 37–38. [Google Scholar]
- Woolfson, D.N.; Bartlett, G.J.; Bruning, M.; Thomson, A.R. New currency for old rope: From coiled-coil assemblies to α-helical barrels. Curr. Opin. Struct. Biol. 2012, 22, 432–441. [Google Scholar] [CrossRef]
- Apostolovic, B.; Danial, M.; Klok, H.-A. Coiled coils: Attractive protein folding motifs for the fabrication of self-assembled, responsive and bioactive materials. Chem. Soc. Rev. 2010, 39, 3541–3575. [Google Scholar] [CrossRef]
- Grigoryan, G.; Keating, A.E. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 2008, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Wood, C.W.; Woolfson, D.N. C CB uilder 2.0: Powerful and accessible coiled-coil modeling. Protein Sci. 2018, 27, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Moll, J.R.; Ruvinov, S.B.; Pastan, I.; Vinson, C. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10−15 M. Protein Sci. 2001, 10, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Gradišar, H.; Jerala, R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J. Pept. Sci. 2011, 17, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.M.; Boyle, A.L.; Bruning, M.; Bartlett, G.J.; Vincent, T.L.; Zaccai, N.R.; Armstrong, C.T.; Bromley, E.H.C.; Booth, P.J.; Brady, R.L. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 2012, 1, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, G.; Reinke, A.W.; Keating, A.E. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature 2009, 458, 859–864. [Google Scholar] [CrossRef]
- Reinke, A.W.; Grant, R.A.; Keating, A.E. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J. Am. Chem. Soc. 2010, 132, 6025–6031. [Google Scholar] [CrossRef] [Green Version]
- Thompson, K.E.; Bashor, C.J.; Lim, W.A.; Keating, A.E. SYNZIP protein interaction toolbox: In vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth. Biol. 2012, 1, 118–129. [Google Scholar] [CrossRef]
- Negron, C.; Keating, A.E. A set of computationally designed orthogonal antiparallel homodimers that expands the synthetic coiled-coil toolkit. J. Am. Chem. Soc. 2014, 136, 16544–16556. [Google Scholar] [CrossRef] [Green Version]
- Crooks, R.O.; Baxter, D.; Panek, A.S.; Lubben, A.T.; Mason, J.M. Deriving heterospecific self-assembling protein–protein interactions using a computational interactome screen. J. Mol. Biol. 2016, 428, 385–398. [Google Scholar] [CrossRef]
- Crooks, R.O.; Lathbridge, A.; Panek, A.S.; Mason, J.M. Computational Prediction and Design for Creating Iteratively Larger Heterospecific Coiled Coil Sets. Biochemistry 2017, 56, 1573–1584. [Google Scholar] [CrossRef] [Green Version]
- Thomson, A.R.; Wood, C.W.; Burton, A.J.; Bartlett, G.J.; Sessions, R.B.; Brady, R.L.; Woolfson, D.N. Computational design of water-soluble α-helical barrels. Science 2014, 346, 485–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, J.B.; Reinke, A.W.; Keating, A.E. Increasing the affinity of selective bZIP-binding peptides through surface residue redesign. Protein Sci. 2014, 23, 940–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drobnak, I.; Gradišar, H.; Ljubetič, A.; Merljak, E.; Jerala, R. Modulation of coiled-coil dimer stability through surface residues while preserving pairing specificity. J. Am. Chem. Soc. 2017, 139, 8229–8236. [Google Scholar] [CrossRef] [PubMed]
- Robson Marsden, H.; Kros, A. Self-assembly of coiled coils in synthetic biology: Inspiration and progress. Angew. Chem. Int. Ed. 2010, 49, 2988–3005. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Collier, J.H. α-Helical coiled-coil peptide materials for biomedical applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1424. [Google Scholar] [CrossRef] [Green Version]
- Pandya, M.J.; Spooner, G.M.; Sunde, M.; Thorpe, J.R.; Rodger, A.; Woolfson, D.N. Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry 2000, 39, 8728–8734. [Google Scholar] [CrossRef]
- Papapostolou, D.; Smith, A.M.; Atkins, E.D.T.; Oliver, S.J.; Ryadnov, M.G.; Serpell, L.C.; Woolfson, D.N. Engineering nanoscale order into a designed protein fiber. Proc. Natl. Acad. Sci. USA 2007, 104, 10853–10858. [Google Scholar] [CrossRef] [Green Version]
- Burgess, N.C.; Sharp, T.H.; Thomas, F.; Wood, C.W.; Thomson, A.R.; Zaccai, N.R.; Brady, R.L.; Serpell, L.C.; Woolfson, D.N. Modular design of self-assembling peptide-based nanotubes. J. Am. Chem. Soc. 2015, 137, 10554–10562. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Liu, R.; Mehta, A.K.; Guerrero-Ferreira, R.C.; Wright, E.R.; Dunin-Horkawicz, S.; Morris, K.; Serpell, L.C.; Zuo, X.; Wall, J.S. Rational design of helical nanotubes from self-assembly of coiled-coil lock washers. J. Am. Chem. Soc. 2013, 135, 15565–15578. [Google Scholar] [CrossRef]
- Park, W.M.; Bedewy, M.; Berggren, K.K.; Keating, A.E. Modular assembly of a protein nanotriangle using orthogonally interacting coiled coils. Sci. Rep. 2017, 7, 10577. [Google Scholar] [CrossRef] [Green Version]
- Boyle, A.L.; Bromley, E.H.C.; Bartlett, G.J.; Sessions, R.B.; Sharp, T.H.; Williams, C.L.; Curmi, P.M.G.; Forde, N.R.; Linke, H.; Woolfson, D.N. Squaring the circle in peptide assembly: From fibers to discrete nanostructures by de novo design. J. Am. Chem. Soc. 2012, 134, 15457–15467. [Google Scholar] [CrossRef] [PubMed]
- Gradišar, H.; Božič, S.; Doles, T.; Vengust, D.; Hafner-Bratkovič, I.; Mertelj, A.; Webb, B.; Šali, A.; Klavžar, S.; Jerala, R. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 2013, 9, 362–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljubetič, A.; Lapenta, F.; Gradišar, H.; Drobnak, I.; Aupič, J.; Strmšek, Ž.; Lainšček, D.; Hafner-Bratkovič, I.; Majerle, A.; Krivec, N. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat. Biotechnol. 2017, 35, 1094–1101. [Google Scholar] [CrossRef] [Green Version]
- Yeates, T.O.; Liu, Y.; Laniado, J. The design of symmetric protein nanomaterials comes of age in theory and practice. Curr. Opin. Struct. Biol. 2016, 39, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, J.M.; Harniman, R.L.; Barnes, F.R.H.; Boyle, A.L.; Collins, A.; Mantell, J.; Sharp, T.H.; Antognozzi, M.; Booth, P.J.; Linden, N. Self-assembling cages from coiled-coil peptide modules. Science 2013, 340, 595–599. [Google Scholar] [CrossRef]
- Raman, S.; Machaidze, G.; Lustig, A.; Aebi, U.; Burkhard, P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 95–102. [Google Scholar] [CrossRef]
- Sciore, A.; Su, M.; Koldewey, P.; Eschweiler, J.D.; Diffley, K.A.; Linhares, B.M.; Ruotolo, B.T.; Bardwell, J.C.A.; Skiniotis, G.; Marsh, E.N.G. Flexible, symmetry-directed approach to assembling protein cages. Proc. Natl. Acad. Sci. USA 2016, 113, 8681–8686. [Google Scholar] [CrossRef] [Green Version]
- Park, W.M.; Champion, J.A. Thermally triggered self-assembly of folded proteins into vesicles. J. Am. Chem. Soc. 2014, 136, 17906–17909. [Google Scholar] [CrossRef]
- Paloni, J.M.; Olsen, B.D. Coiled-Coil Domains for Self-Assembly and Sensitivity Enhancement of Protein–Polymer Conjugate Biosensors. ACS Appl. Polym. Mater. 2020, 2, 1114–1123. [Google Scholar] [CrossRef]
- Xu, C.; Kopeček, J. Genetically engineered block copolymers: Influence of the length and structure of the coiled-coil blocks on hydrogel self-assembly. Pharm. Res. 2008, 25, 674–682. [Google Scholar] [CrossRef]
- Nambiar, M.; Nepal, M.; Chmielewski, J. Self-Assembling Coiled-Coil Peptide Nanotubes with Biomolecular Cargo Encapsulation. ACS Biomater. Sci. Eng. 2019, 5, 5082–5087. [Google Scholar] [CrossRef]
- Ross, J.F.; Bridges, A.; Fletcher, J.M.; Shoemark, D.; Alibhai, D.; Bray, H.E.V.; Beesley, J.L.; Dawson, W.M.; Hodgson, L.R.; Mantell, J.; et al. Decorating Self-Assembled Peptide Cages with Proteins. ACS Nano 2017, 11, 7901–7914. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Choi, W.T.; Heller, W.T.; Ke, Z.; Wright, E.R.; Champion, J.A. Engineering globular protein vesicles through tunable self-assembly of recombinant fusion proteins. Small 2017, 13, 1700399. [Google Scholar] [CrossRef] [PubMed]
- Cristie-David, A.S.; Chen, J.; Nowak, D.B.; Bondy, A.L.; Sun, K.; Park, S.I.; Banaszak Holl, M.M.; Su, M.; Marsh, E.N.G. Coiled-coil-mediated assembly of an icosahedral protein cage with extremely high thermal and chemical stability. J. Am. Chem. Soc. 2019, 141, 9207–9216. [Google Scholar] [CrossRef] [PubMed]
- Cristie-David, A.S.; Koldewey, P.; Meinen, B.A.; Bardwell, J.C.A.; Marsh, E.N.G. Elaborating a coiled-coil-assembled octahedral protein cage with additional protein domains. Protein Sci. 2018, 27, 1893–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petka, W.A.; Harden, J.L.; McGrath, K.P.; Wirtz, D.; Tirrell, D.A. Reversible hydrogels from self-assembling artificial proteins. Science 1998, 281, 389–392. [Google Scholar] [CrossRef]
- Xu, C.; Breedveld, V.; Kopeček, J. Reversible Hydrogels from Self-Assembling Genetically Engineered Protein Block Copolymers. Biomacromolecules 2005, 6, 1739–1749. [Google Scholar] [CrossRef]
- Banwell, E.F.; Abelardo, E.S.; Adams, D.J.; Birchall, M.A.; Corrigan, A.; Donald, A.M.; Kirkland, M.; Serpell, L.C.; Butler, M.F.; Woolfson, D.N. Rational design and application of responsive α-helical peptide hydrogels. Nat. Mater. 2009, 8, 596–600. [Google Scholar] [CrossRef] [Green Version]
- Thomas, F.; Burgess, N.C.; Thomson, A.R.; Woolfson, D.N. Controlling the assembly of coiled–coil peptide nanotubes. Angew. Chem. Int. Ed. 2016, 55, 987–991. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; An, B.; Bagheri, M.; Wang, Q.; Harden, J.L.; Kaplan, D.L. Electrochemically Directed Assembly of Designer Coiled-Coil Telechelic Proteins. ACS Biomater. Sci. Eng. 2017, 3, 3195–3206. [Google Scholar] [CrossRef]
- Shen, W.; Zhang, K.; Kornfield, J.A.; Tirrell, D.A. Tuning the erosion rate of artificial protein hydrogels through control of network topology. Nat. Mater. 2006, 5, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Dooling, L.J.; Buck, M.E.; Zhang, W.-B.; Tirrell, D.A. Programming Molecular Association and Viscoelastic Behavior in Protein Networks. Adv. Mater. 2016, 28, 4651–4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dooling, L.J.; Tirrell, D.A. Engineering the Dynamic Properties of Protein Networks through Sequence Variation. ACS Cent. Sci. 2016, 2, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.K.; Wong, K.M.; Lee, E.J.; Le, M.M.; Patel, D.M.; Paravastu, A.K. Post-assembly α-helix to β-sheet structural transformation within SAF-p1/p2a peptide nanofibers. Soft Matter 2018, 14, 8986–8996. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.S.; Glassman, M.J.; Olsen, B.D. Solid-state nanostructured materials from self-assembly of a globular protein–polymer diblock copolymer. ACS Nano 2011, 5, 5697–5707. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Obermeyer, A.C.; Olsen, B.D. Three-Dimensional Ordered Antibody Arrays Through Self-Assembly of Antibody–Polymer Conjugates. Angew. Chem. Int. Ed. 2017, 56, 1273–1277. [Google Scholar] [CrossRef]
- Huang, A.; Paloni, J.M.; Wang, A.; Obermeyer, A.C.; Sureka, H.V.; Yao, H.; Olsen, B.D. Predicting Protein–Polymer Block Copolymer Self-Assembly from Protein Properties. Biomacromolecules 2019, 20, 3713–3723. [Google Scholar] [CrossRef] [Green Version]
- Nepal, M.; Sheedlo, M.J.; Das, C.; Chmielewski, J. Accessing three-dimensional crystals with incorporated guests through metal-directed coiled-coil peptide assembly. J. Am. Chem. Soc. 2016, 138, 11051–11057. [Google Scholar] [CrossRef]
- Galloway, J.M.; Senior, L.; Fletcher, J.M.; Beesley, J.L.; Hodgson, L.R.; Harniman, R.L.; Mantell, J.M.; Coombs, J.; Rhys, G.G.; Xue, W.-F. Bioinspired Silicification Reveals Structural Detail in Self-Assembled Peptide Cages. ACS Nano 2017, 12, 1420–1432. [Google Scholar] [CrossRef]
- Mosayebi, M.; Shoemark, D.K.; Fletcher, J.M.; Sessions, R.B.; Linden, N.; Woolfson, D.N.; Liverpool, T.B. Beyond icosahedral symmetry in packings of proteins in spherical shells. Proc. Natl. Acad. Sci. USA 2017, 114, 9014–9019. [Google Scholar] [CrossRef] [Green Version]
- Beesley, J.L.; Baum, H.E.; Hodgson, L.R.; Verkade, P.; Banting, G.S.; Woolfson, D.N. Modifying self-assembled peptide cages to control internalization into mammalian cells. Nano Lett. 2018, 18, 5933–5937. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Glennie, S.J.; Lam, H.S.; Baum, H.E.; Kandage, D.; Williams, N.A.; Morgan, D.J.; Woolfson, D.N.; Davidson, A.D. A Modular Vaccine Platform Combining Self-Assembled Peptide Cages and Immunogenic Peptides. Adv. Funct. Mater. 2019, 29, 1807357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indelicato, G.; Wahome, N.; Ringler, P.; Müller, S.A.; Nieh, M.-P.; Burkhard, P.; Twarock, R. Principles governing the self-assembly of coiled-coil protein nanoparticles. Biophys. J. 2016, 110, 646–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, U.; Graff, A.; Buchmeier, S.; Rigler, P.; Silvan, U.; Tropel, D.; Jockusch, B.M.; Aebi, U.; Burkhard, P.; Schoenenberger, C.-A. Peptide nanoparticles serve as a powerful platform for the immunogenic display of poorly antigenic actin determinants. J. Mol. Biol. 2009, 386, 1368–1381. [Google Scholar] [CrossRef] [PubMed]
- IYang, Y.; Ringler, P.; Müller, S.A.; Burkhard, P. Optimizing the refolding conditions of self-assembling polypeptide nanoparticles that serve as repetitive antigen display systems. J. Struct. Biol. 2012, 177, 168–176. [Google Scholar] [CrossRef]
- Kaba, S.A.; Brando, C.; Guo, Q.; Mittelholzer, C.; Raman, S.; Tropel, D.; Aebi, U.; Burkhard, P.; Lanar, D.E. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J. Immunol. 2009, 183, 7268–7277. [Google Scholar] [CrossRef] [Green Version]
- Karch, C.P.; Li, J.; Kulangara, C.; Paulillo, S.M.; Raman, S.K.; Emadi, S.; Tan, A.; Helal, Z.H.; Fan, Q.; Khan, M.I. Vaccination with self-adjuvanted protein nanoparticles provides protection against lethal influenza challenge. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 241–251. [Google Scholar] [CrossRef]
- Karch, C.P.; Bai, H.; Torres, O.B.; Tucker, C.A.; Michael, N.L.; Matyas, G.R.; Rolland, M.; Burkhard, P.; Beck, Z. Design and characterization of a self-assembling protein nanoparticle displaying HIV-1 Env V1V2 loop in a native-like trimeric conformation as vaccine antigen. Nanomed. Nanotechnol. Biol. Med. 2019, 16, 206–216. [Google Scholar] [CrossRef]
- Wahome, N.; Pfeiffer, T.; Ambiel, I.; Yang, Y.; Keppler, O.T.; Bosch, V.; Burkhard, P. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chem. Biol. Drug Des. 2012, 80, 349–357. [Google Scholar] [CrossRef]
- Guo, Q.; Dasgupta, D.; Doll, T.A.P.F.; Burkhard, P.; Lanar, D.E. Expression, purification and refolding of a self-assembling protein nanoparticle (SAPN) malaria vaccine. Methods 2013, 60, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Badieyan, S.; Sciore, A.; Eschweiler, J.D.; Koldewey, P.; Cristie-David, A.S.; Ruotolo, B.T.; Bardwell, J.C.A.; Su, M.; Marsh, E.N.G. Symmetry-directed self-assembly of a tetrahedral protein cage mediated by de novo-designed coiled coils. ChemBioChem 2017, 18, 1888–1892. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapenta, F.; Aupič, J.; Strmšek, Ž.; Jerala, R. Coiled coil protein origami: From modular design principles towards biotechnological applications. Chem. Soc. Rev. 2018, 47, 3530–3542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristie-David, A.S.; Sciore, A.; Badieyan, S.; Escheweiler, J.D.; Koldewey, P.; Bardwell, J.C.A.; Ruotolo, B.T.; Marsh, E.N.G. Evaluation of de novo-designed coiled coils as off-the-shelf components for protein assembly. Mol. Syst. Des. Eng. 2017, 2, 140–148. [Google Scholar] [CrossRef]
- DeFrates, K.; Markiewicz, T.; Gallo, P.; Rack, A.; Weyhmiller, A.; Jarmusik, B.; Hu, X. Protein polymer-based nanoparticles: Fabrication and medical applications. Int. J. Mol. Sci. 2018, 19, 1717. [Google Scholar] [CrossRef] [Green Version]
- Park, W.M.; Champion, J.A. Two-Step Protein Self-Assembly in the Extracellular Matrix. Angew. Chem. Int. Ed. 2013, 52, 8098–8101. [Google Scholar] [CrossRef]
- Park, W.M.; Yee, C.M.; Champion, J.A. Self-assembled hybrid supraparticles that proteolytically degrade tumor necrosis factor-α. J. Mater. Chem. B 2016, 4, 1633–1639. [Google Scholar] [CrossRef]
- Caparco, A.A.; Bommarius, B.R.; Bommarius, A.S.; Champion, J.A. Protein-inorganic calcium-phosphate supraparticles as a robust platform for enzyme co-immobilization. Biotechnol. Bioeng. 2020. [Google Scholar] [CrossRef]
- Simnick, A.J.; Valencia, C.A.; Liu, R.; Chilkoti, A. Morphing low-affinity ligands into high-avidity nanoparticles by thermally triggered self-assembly of a genetically encoded polymer. ACS Nano 2010, 4, 2217–2227. [Google Scholar] [CrossRef] [Green Version]
- Diehl, M.R.; Zhang, K.; Lee, H.J.; Tirrell, D.A. Engineering cooperativity in biomotor-protein assemblies. Science 2006, 311, 1468–1471. [Google Scholar] [CrossRef] [Green Version]
- Shemesh, O.A.; Linghu, C.; Piatkevich, K.D.; Goodwin, D.; Gritton, H.; Romano, M.F.; Siciliano, C.A.; Gao, R.; Yu, C.-C.J.; Tseng, H. Precision calcium imaging of dense neural populations via a cell body-targeted calcium indicator. bioRxiv 2019, 773069. [Google Scholar]
- Park, W.M.; Champion, J.A. Colloidal Assembly of Hierarchically Structured Porous Supraparticles from Flower-Shaped Protein–Inorganic Hybrid Nanoparticles. ACS Nano 2016, 10, 8271–8280. [Google Scholar] [CrossRef] [PubMed]





| Category | Type | Length Scale 1 | Design Strategy | References |
|---|---|---|---|---|
| Unbounded nanostructures | Network | ~500 nm ~50–100 nm | Coiled-coil fusion Modification | [46,47,48] |
| Fiber | ~50–100 nm | Coiled-coil fusion/Modification | [26,27] | |
| Tube | ~3–100 nm | Modification | [28,49] | |
| Ordered array | ~25 nm | Fusion to other domains | [39] | |
| Discrete nanoparticles | Cage | ~100 nm | Coiled-coil conjugation | [35] |
| Vesicle | ~1–2 µm | Fusion to other domains | [38] | |
| Polyhedron | ~16–24 nm~18–25 nm | Coiled-coil fusion Fusion to other domains | [36,37,44] | |
| Origami nanostructures | Polyhedron | ~5–10 nm | Single-chain CCPO | [32,33] |
| Polygon | ~5–10 nm | Multi-chain CCPO | [30,31] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, W.M. Coiled-Coils: The Molecular Zippers that Self-Assemble Protein Nanostructures. Int. J. Mol. Sci. 2020, 21, 3584. https://doi.org/10.3390/ijms21103584
Park WM. Coiled-Coils: The Molecular Zippers that Self-Assemble Protein Nanostructures. International Journal of Molecular Sciences. 2020; 21(10):3584. https://doi.org/10.3390/ijms21103584
Chicago/Turabian StylePark, Won Min. 2020. "Coiled-Coils: The Molecular Zippers that Self-Assemble Protein Nanostructures" International Journal of Molecular Sciences 21, no. 10: 3584. https://doi.org/10.3390/ijms21103584