Transforming Growth Factor-β Promotes Morphomechanical Effects Involved in Epithelial to Mesenchymal Transition in Living Hepatocellular Carcinoma
Abstract
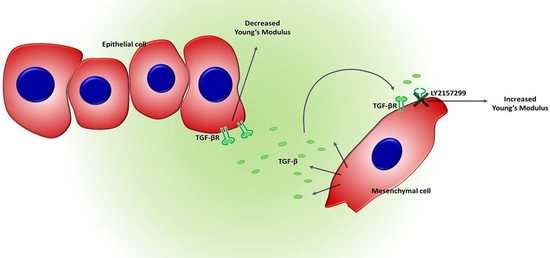
:1. Introduction
- -
- Hep3B and PLC/PRF/5 cell lines, having an epithelial-like phenotype. These cell lines do not secrete TGF-β1.
- -
- HLE cell line, having a mesenchymal-like phenotype. These cells produce autocrine TGF-β1.
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Samples
4.3. Instruments
4.4. AFM Experiments
4.5. Confocal Experiments
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bao, G.; Suresh, S. Cell and molecular mechanics of biological materials. Nat. Mater. 2003, 2, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kamm, R.D.; Lee, R.T. Cell mechanics and mechanotransduction: Pathways, probes, and physiology. Am. J. Physiol. Cell Physiol. 2004, 287, C1–C11. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Koshy, S.T.; Branco da Cunha, C.; Shin, J.W.; Verbeke, C.S.; Allison, K.H.; Mooney, D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014, 13, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Xia, Y.; Discher, D.E.; Janmey, P.A. Mechanotransduction in cancer. Curr. Opin. Chem. Eng. 2016, 11, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.S.; Lee, G.Y.; Ong, C.N.; Lim, C.T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.M.; Zauscher, S.; Guilak, F. Viscoelastic properties of zonal articular chondrocytes measured by atomic force microscopy. Osteoarthr. Cartil. 2006, 14, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.M.; Topel, M.; Zauscher, S.; Vail, T.P.; Guilak, F. Viscoelastic properties of human mesenchymally-derived stem cells and primary osteoblasts, chondrocytes, and adipocytes. J. Biomech. 2008, 41, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Hay, E.D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995, 154, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007, 98, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, J.P. The lymph node as a bridgehead in the metastatic dissemination of tumors. Recent Res. Cancer 2000, 157, 55–81. [Google Scholar]
- Nagaraj, N.S.; Datta, P.K. Targeting the transforming growth factor-β signaling pathway in human cancer. Expert Opin. Investig. Drugs 2010, 19, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the epithelial-mesenchymal transition by TGF-β. Future Oncol. 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Boil. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.; Kale, V.P. TGF-β signaling and its role in the regulation of hematopoietic stem cells. Syst. Synth. Boil. 2015, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Hajjar, D.P.; Tauras, J.M.; Feng, J.; Gotto, A.M., Jr.; Nicholson, A.C. Transforming growth factor-β1 (TGF-β1) and TGF-β2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-γ. J. Boil. Chem. 2000, 275, 1241–1246. [Google Scholar] [CrossRef]
- Massague, J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Boil. 2000, 1, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.L.; Blobe, G.C. Role of transforming growth factor β in human cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 2078–2093. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Katsuno, Y.; Lamouille, S.; Derynck, R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013, 25, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Herbertz, S.; Sawyer, J.S.; Stauber, A.J.; Gueorguieva, I.; Driscoll, K.E.; Estrem, S.T.; Cleverly, A.L.; Desaiah, D.; Guba, S.C.; Benhadji, K.A.; et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-β signaling pathway. Drug Des. Dev. Ther. 2015, 9, 4479–4499. [Google Scholar]
- Giannelli, G.; Mazzocca, A.; Fransvea, E.; Lahn, M.; Antonaci, S. Inhibiting TGF-β signaling in hepatocellular carcinoma. Biochim. Biophys. Acta 2011, 1815, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, Y.; Nokihara, H.; Yamada, Y.; Yamamoto, N.; Sunami, K.; Utsumi, H.; Asou, H.; Takahash, I.O.; Ogasawara, K.; Gueorguieva, I.; et al. Phase 1 study of galunisertib, a TGF-β receptor I kinase inhibitor, in Japanese patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2015, 76, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Dituri, F.; Serio, G.; Filannino, D.; Mascolo, A.; Sacco, R.; Villa, E.; Giannelli, G. Circulating TGF-β1-related biomarkers in patients with hepatocellular carcinoma and their association with HCC staging scores. Cancer Lett. 2014, 353, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Dzieran, J.; Fabian, J.; Feng, T.; Coulouarn, C.; Ilkavets, I.; Kyselova, A.; Breuhahn, K.; Dooley, S.; Meindl-Beinker, N.M. Comparative analysis of TGF-β/Smad signaling dependent cytostasis in human hepatocellular carcinoma cell lines. PLoS ONE 2013, 8, e72252. [Google Scholar] [CrossRef] [PubMed]
- Serova, M.; Tijeras-Raballand, A.; Dos Santos, C.; Albuquerque, M.; Paradis, V.; Neuzillet, C.; Benhadji, K.A.; Raymond, E.; Faivre, S.; de Gramont, A. Effects of TGF-β signalling inhibition with galunisertib (LY2157299) in hepatocellular carcinoma models and in ex vivo whole tumor tissue samples from patients. Oncotarget 2015, 6, 21614–21627. [Google Scholar] [CrossRef] [PubMed]
- Sheetz, M.P. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Boil. 2001, 2, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Cascione, M.; de Matteis, V.; Rinaldi, R.; Leporatti, S. Atomic force microscopy combined with optical microscopy for cells investigation. Microsc. Res. Tech. 2017, 80, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Thoelking, G.; Reiss, B.; Wegener, J.; Oberleithner, H.; Pavenstaedt, H.; Riethmuller, C. Nanotopography follows force in TGF-β1 stimulated epithelium. Nanotechnology 2010, 21, 265102. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.T.; Medina, C.; Davies, A.M.; Ehrhardt, C. Cytoskeletal re-arrangement in TGF-β1-induced alveolar epithelial-mesenchymal transition studied by atomic force microscopy and high-content analysis. Nanomed. Nanotechnol. Boil. Med. 2012, 8, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Baronsky, T.; Pietuch, A.; Rother, J.; Oelkers, M.; Fichtner, D.; Wedlich, D.; Janshoff, A. Tension monitoring during epithelial-to-mesenchymal transition links the switch of phenotype to expression of moesin and cadherins in NMuMG cells. PLoS ONE 2013, 8, e80068. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Hong, J.W.; Kim, J.; Shin, J.H. Comparative study on the differential mechanical properties of human liver cancer and normal cells. Anim. Cells Syst. 2013, 17, 170–178. [Google Scholar] [CrossRef] [Green Version]
- Stylianou, A.; Gkretsi, V.; Stylianopoulos, T. Transforming growth factor-β modulates pancreatic cancer associated fibroblasts cell shape, stiffness and invasion. Biochim. Biophys. Acta 2018, 1862, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Cascione, M.; De Matteis, V.; Toma, C.C.; Leporatti, S. Morphomechanical Alterations Induced by Transforming Growth Factor-β1 in Epithelial Breast Cancer Cells. Cancers 2018, 10, 234. [Google Scholar] [CrossRef] [PubMed]
- Malfettone, A.; Soukupova, J.; Bertran, E.; Crosas-Molist, E.; Lastra, R.; Fernando, J.; Koudelkova, P.; Rani, B.; Fabra, A.; Serrano, T.; et al. Transforming growth factor-β-induced plasticity causes a migratory stemness phenotype in hepatocellular carcinoma. Cancer Lett. 2017, 392, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Weichsel, J.; Herold, N.; Lehmann, M.J.; Krausslich, H.G.; Schwarz, U.S. A Quantitative Measure for Alterations in the Actin Cytoskeleton Investigated with Automated High-Throughput Microscopy. Cytom. Part A 2010, 77, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Jahne, B. Digital Image Processing; Springer: Berlin, Germany, 2005; Volume 4. [Google Scholar]
- Zhang, W.; Kai, K.; Ueno, N.T.; Qin, L. A Brief Review of the Biophysical Hallmarks of Metastatic Cancer Cells. Cancer Hallm. 2013, 1, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mierke, C.T. The fundamental role of mechanical properties in the progression of cancer disease and inflammation. Rep. Prog. Phys. Phys. Soc. 2014, 77, 076602. [Google Scholar] [CrossRef] [PubMed]
- Alibert, C.; Goud, B.; Manneville, J.B. Are cancer cells really softer than normal cells? Boil. Cell 2017, 109, 167–189. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Moreno-Caceres, J.; Sanchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P.; Consortium, I.-L. TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Villa, E.; Lahn, M. Transforming growth factor-β as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014, 74, 1890–1894. [Google Scholar] [CrossRef] [PubMed]
- Filippi-Chiela, E.C.; Oliveira, M.M.; Jurkovski, B.; Callegari-Jacques, S.M.; da Silva, V.D.; Lenz, G. Nuclear morphometric analysis (NMA): Screening of senescence, apoptosis and nuclear irregularities. PLoS ONE 2012, 7, e42522. [Google Scholar] [CrossRef] [PubMed]
- Zdilla, M.J.; Hatfield, S.A.; McLean, K.A.; Cyrus, L.M.; Laslo, J.M.; Lambert, H.W. Circularity, Solidity, Axes of a Best Fit Ellipse, Aspect Ratio, and Roundness of the Foramen Ovale: A Morphometric Analysis with Neurosurgical Considerations. J. Craniofac. Surg. 2016, 27, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Kole, T.P.; Tseng, Y.; Huang, L.; Katz, J.L.; Wirtz, D. Rho kinase regulates the intracellular micromechanical response of adherent cells to rho activation. Mol. Boil. Cell 2004, 15, 3475–3484. [Google Scholar] [CrossRef] [PubMed]
- Cascione, M.; de Matteis, V.; Toma, C.C.; Pellegrino, P.; Leporatti, S.; Rinaldi, R. Morphomechanical and structural changes induced by ROCK inhibitor in breast cancer cells. Exp. Cell Res. 2017, 360, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, V.; Lucchetti, D.; Maiorana, A.; Papi, M.; Maulucci, G.; Calapa, F.; Ciasca, G.; Giordano, R.; Sgambato, A.; de Spirito, M. Mechanical and structural comparison between primary tumor and lymph node metastasis cells in colorectal cancer. Soft Matter 2015, 11, 5719–5726. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA A Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Dituri, F.; Mazzocca, A.; Fernando, J.; Papappicco, P.; Fabregat, I.; de Santis, F.; Paradiso, A.; Sabba, C.; Giannelli, G. Differential Inhibition of the TGF-β Signaling Pathway in HCC Cells Using the Small Molecule Inhibitor LY2157299 and the D10 Monoclonal Antibody against TGF-β Receptor Type II. PLoS ONE 2013, 8, e67109. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K.; Lammerding, J. Nuclear mechanics during cell migration. Curr. Opin. Cell Boil. 2011, 23, 55–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, P. Cancer nucleus: Morphology and beyond. Diagn. Cytopathol. 2010, 38, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.J.; Jaschke, M. Calculation of Thermal Noise in Atomic-Force Microscopy. Nanotechnology 1995, 6, 1–7. [Google Scholar] [CrossRef]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cascione, M.; Leporatti, S.; Dituri, F.; Giannelli, G. Transforming Growth Factor-β Promotes Morphomechanical Effects Involved in Epithelial to Mesenchymal Transition in Living Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 108. https://doi.org/10.3390/ijms20010108
Cascione M, Leporatti S, Dituri F, Giannelli G. Transforming Growth Factor-β Promotes Morphomechanical Effects Involved in Epithelial to Mesenchymal Transition in Living Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2019; 20(1):108. https://doi.org/10.3390/ijms20010108
Chicago/Turabian StyleCascione, Mariafrancesca, Stefano Leporatti, Francesco Dituri, and Gianluigi Giannelli. 2019. "Transforming Growth Factor-β Promotes Morphomechanical Effects Involved in Epithelial to Mesenchymal Transition in Living Hepatocellular Carcinoma" International Journal of Molecular Sciences 20, no. 1: 108. https://doi.org/10.3390/ijms20010108