Thymoquinone Suppresses IRF-3-Mediated Expression of Type I Interferons via Suppression of TBK1
Abstract
:1. Introduction
2. Results
2.1. Effect of Thymoquinone on the mRNA Expression of Interferon Genes in LPS-Stimulated RAW264.7 Cells
2.2. Effect of Thymoquinone on the Transcriptional Activation of IRF-3
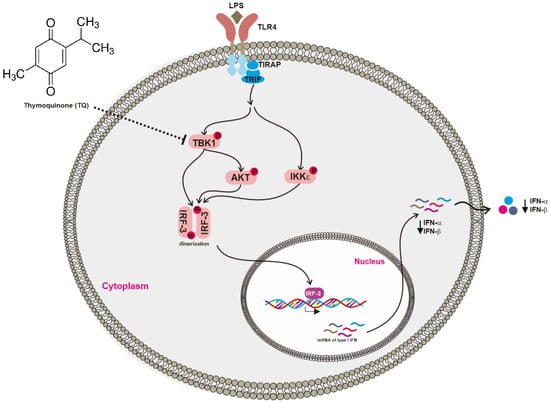
2.3. Effect of Thymoquinone on Upstream Signaling Events Regulating IRF-3 Activation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Drug Preparation
4.3. Cell Viability Assay
4.4. mRNA Analysis by Semiquantitative Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR) and Real-Time PCR (qPCR)
4.5. Luciferase Reporter Gene Assay
4.6. Preparation of Whole Cell Lysates and Immunoblotting
4.7. Cellular Thermal Shift Assay (CETSA)
4.8. Statistical Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern Recognition Receptors and the Innate Immune Response to Viral Infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [PubMed]
- Vidya, M.K.; Kumar, V.G.; Sejian, V.; Bagath, M.; Krishnan, G.; Bhatta, R. Toll-like receptors: Significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 2017, 37, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.; Mages, J.; Heit, A.; Lang, R.; Wagner, H. Transcriptional activation induced in macrophages by Toll-like receptor (TLR) ligands: From expression profiling to a model of TLR signaling. Eur. J. Immunol. 2004, 34, 2863–2873. [Google Scholar] [CrossRef] [PubMed]
- Dragan, A.I.; Hargreaves, V.V.; Makeyeva, E.N.; Privalov, P.L. Mechanisms of activation of interferon regulator factor 3: The role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res. 2007, 35, 3525–3534. [Google Scholar] [CrossRef] [PubMed]
- Barnes, B.; Lubyova, B.; Pitha, P.M. On the role of IRF in host defense. J. Interferon Cytokine Res. 2002, 22, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007, 282, 15325–15329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, M.; Chen, L.; Yang, K.; Shan, Y.; Zhu, L.; Sun, S.; Li, L.; Wang, C. The TAK1-JNK cascade is required for IRF3 function in the innate immune response. Cell. Res. 2009, 19, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, S.; Taylor, J.; Strickson, S.; McCartney, T.; Cohen, P. Identification of TBK1 complexes required for the phosphorylation of IRF3 and the production of interferon beta. Biochem. J. 2017, 474, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.J.; Yang, W.S.; Kim, D.; Aravinthan, A.; Kim, J.H.; Cho, J.Y. Thymoquinone: An IRAK1 inhibitor with in vivo and in vitro anti-inflammatory activities. Sci. Rep. 2017, 7, 42995. [Google Scholar] [CrossRef] [PubMed]
- Khader, M.; Eckl, P.M. Thymoquinone: An emerging natural drug with a wide range of medical applications. Iran. J. Basic Med. Sci. 2014, 17, 950–957. [Google Scholar] [PubMed]
- Wang, Z.; Ji, J.; Peng, D.; Ma, F.; Cheng, G.; Qin, F.X.-F. Complex regulation pattern of IRF3 activation revealed by a novel dimerization reporter system. J. Immunol. 2016, 196, 4322–4330. [Google Scholar] [CrossRef] [PubMed]
- McWhirter, S.M.; Fitzgerald, K.A.; Rosains, J.; Rowe, D.C.; Golenbock, D.T.; Maniatis, T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 2004, 101, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I Inteferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- McCoy, C.E.; Carpenter, S.; Palsson-McDermott, E.M.; Gearing, L.J.; O’Neill, L.A. Glucocorticoids inhibit IRF3 phosphorylation in response to Toll-like receptor-3 and -4 by targeting TBK1 activation. J. Biol. Chem. 2008, 283, 14277–14285. [Google Scholar] [CrossRef] [PubMed]
- Servant, M.J.; Grandvaux, N.; Benjamin, R.; Duguay, D.; Lin, R.; Hiscott, J. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 2003, 278, 9441–9447. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.-T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. TLR pathways and IFN-regulatory factors: To each its own. Eur. J. Immunol. 2007, 37, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Joung, S.M.; Park, Z.-Y.; Rani, S.; Takeuchi, O.; Akira, S.; Lee, J.Y. Akt Contributes to Activation of the TRIF-Dependent Signaling Pathways of TLRs by Interacting with TANK-Binding Kinase 1. J. Immunol. 2011, 186, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, E.; Berghe, W.V.; Detienne, S.; Amraoui, Z.; Fitzgerald, K.A.; Haegeman, G.; Goldman, M.; Willems, F. Inhibition of phosphoinositide 3-kinase enhances TRIF-dependent NF-κB activation and IFN-β synthesis downstream of Toll-like receptor 3 and 4. Eur. J. Immunol. 2005, 35, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Plater, L.; Peggie, M.; Cohen, P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 2009, 284, 14136–14146. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Helgason, E.; Phung, Q.T.; Quan, C.L.; Iyer, R.S.; Lee, M.W.; Bowman, K.K.; Starovasnik, M.A.; Dueber, E.C. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. USA 2012, 109, 9378–9383. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Sankaran, B.; Chaton, C.T.; Herr, A.B.; Mishra, A.; Peng, J.; Li, P. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 2013, 21, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Leveridge, M.; Norling, C.; Karén, J.; Molina, D.M.; O’Neill, D.; Dowling, J.E.; Davey, P.; Cowan, S.; Dabrowski, M.; et al. Determining direct binders of the Androgen Receptor using a high-throughput Cellular Thermal Shift Assay. Sci. Rep. 2018, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Helgason, E.; Phung, Q.T.; Dueber, E.C. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013, 587, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Devos, J.M.; Ng, S.-L.; Nanao, M.H.; Round, A.; Maniatis, T.; Panne, D. Crystal structure and mechanism of activation of TANK-Binding Kinase 1. Cell. Rep. 2013, 3, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Chen, Z.J. STING specifies IRF3 phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci. Signal. 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- Han, S.Y.; Kim, J.; Kim, E.; Kim, S.H.; Seo, D.B.; Kim, J.-H.; Shin, S.S.; Cho, J.Y. AKT-targeted anti-inflammatory activity of Panax ginseng calyx ethanolic extract. J. Ginseng Res. 2017, in press. [Google Scholar] [CrossRef]
- Park, J.G.; Son, Y.-J.; Yoo, B.C.; Yang, W.S.; Kim, J.H.; Kim, J.-H.; Cho, J.Y. Syk plays a critical role in the expression and activation of IRAK1 in LPS-treated macrophages. Mediators Inflamm. 2017, 2017, 1506248. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, H.G.; Han, S.Y.; Jeong, D.; Yang, W.S.; Kim, J.-I.; Kim, J.H.; Yi, Y.-S.; Cho, J.Y. Hydroquinone suppresses IFN-β expression by targeting AKT/IRF3 pathway. Korean J. Physiol. Pharmacol. 2017, 21, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.S.; Yi, Y.S.; Son, Y.J.; Jeong, D.; Sung, N.Y.; Aravinthan, A.; Kim, J.H.; Cho, J.Y. Comparison of anticancer activities of Korean Red Ginseng-derived fractions. J. Ginseng Res. 2017, 41, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.J.; Hong, Y.D.; Baek, K.S.; Yoo, S.; Hong, Y.H.; Kim, J.H.; Lee, J.O.; Kim, D.; Park, J.; Cho, J.Y. In vitro antioxidative and anti-inflammatory effects of the compound K-rich fraction BIOGF1K, prepared from Panax ginseng. J. Ginseng Res. 2017, 41, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Yi, Y.S.; Kim, D.; Kim, M.H.; Park, J.G.; Kim, E.; Lee, S.Y.; Yoon, K.; Kim, J.H.; Park, J.; et al. Nuclear factor kappa-B- and activator protein-1-mediated immunostimulatory activity of compound K in monocytes and macrophages. J. Ginseng Res. 2017, 41, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Okai, T.; Iwatani-Yoshihara, M.; Mochizuki, M.; Unno, S.; Kuno, M.; Yoshikawa, M.; Shibata, S.; Nakakariya, M.; Yogo, T.; et al. CETSA quantitatively verifies in vivo target engagement of novel RIPK1 inhibitors in various biospecimens. Sci. Rep. 2017, 7, 13000. [Google Scholar] [CrossRef] [PubMed]






| PCR Type | Name | Sequence (5′–3′) | |
|---|---|---|---|
| Reverse Transcriptase PCR | IFN-α | Forward Reverse | CAACACCTACACAGGTTACC AGTGGCTTCCCAGATGTTCC |
| IFN-β | Forward Reverse | TCCAAGAAAGGACGAACATT TGAGGACATCTCCCACGTCA | |
| GAPDH | Forward Reverse | ACCACAGTCCATGCCATCAC CCACCACCCTGTTGCTGTAG | |
| Real Time PCR | IFN-β | Forward Reverse | AAGAGTTACACTGCCTTTGCTATC CACTGTCTGCTGGTGGAGTTCATC |
| GAPDH | Forward Reverse | CAATGAATACGGCTACAGCA AGGGAGATGCTCAGTGTTGG | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aziz, N.; Son, Y.-J.; Cho, J.Y. Thymoquinone Suppresses IRF-3-Mediated Expression of Type I Interferons via Suppression of TBK1. Int. J. Mol. Sci. 2018, 19, 1355. https://doi.org/10.3390/ijms19051355
Aziz N, Son Y-J, Cho JY. Thymoquinone Suppresses IRF-3-Mediated Expression of Type I Interferons via Suppression of TBK1. International Journal of Molecular Sciences. 2018; 19(5):1355. https://doi.org/10.3390/ijms19051355
Chicago/Turabian StyleAziz, Nur, Young-Jin Son, and Jae Youl Cho. 2018. "Thymoquinone Suppresses IRF-3-Mediated Expression of Type I Interferons via Suppression of TBK1" International Journal of Molecular Sciences 19, no. 5: 1355. https://doi.org/10.3390/ijms19051355