Decitabine and Melphalan Fail to Reactivate p73 in p53 Deficient Myeloma Cells
Abstract
:1. Introduction
2. Results
2.1. TP73 Is Preferentially Constitutively Expressed in TP53wt HMCLs
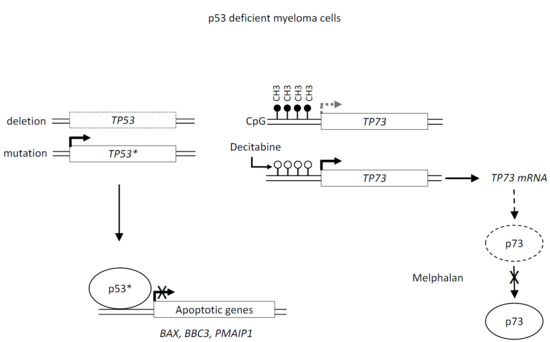
2.2. Decitabine Decreased TP73 Methylation and Induced TP73 Expression
2.3. Melphalan and Cisplatin Induced TP73 Expression in TP53wt HMCLs but Not in Decitabine-Treated TP53mut HMCLs
3. Discussion
4. Materials and Methods
4.1. HMCLs and Reagents
4.2. Bisulfite Treatment, Methylation Specific PCRs and RT-PCR
4.3 Western Blotting
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HMCL | Human Myeloma Cell Line |
| MM | Multiple Myeloma |
| MS-PCR | Methylation-specific PCR |
References
- Fonseca, R.; Bergsagel, P.L.; Drach, J.; Shaughnessy, J.; Gutierrez, N.; Stewart, A.K.; Morgan, G.; van Ness, B.; Chesi, M.; Minvielle, S.; et al. International Myeloma Working, International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia 2009, 23, 2210–2221. [Google Scholar] [CrossRef] [PubMed]
- Avet-Loiseau, H.; Attal, M.; Campion, L.; Caillot, D.; Hulin, C.; Marit, G.; Stoppa, A.M.; Voillat, L.; Wetterwald, M.; Pegourie, B.; et al. Long-term analysis of the IFM 99 trials for myeloma: Cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J. Clin. Oncol. 2012, 30, 1949–1952. [Google Scholar] [CrossRef] [PubMed]
- Lode, L.; Eveillard, M.; Trichet, V.; Soussi, T.; Wuilleme, S.; Richebourg, S.; Magrangeas, F.; Ifrah, N.; Campion, L.; Traulle, C.; et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 2010, 95, 1973–1976. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, N.; Ashby, C.; Rasche, L.; Chavan, S.S.; Stein, C.; Stephens, O.W.; Tytarenko, R.; Bauer, M.A.; Meissner, T.; Deshpande, S.; et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016, 128, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, C.; Jara-Acevedo, M.; Corchete, L.A.; Castillo, D.; Ordonez, G.R.; Sarasquete, M.E.; Puig, N.; Martinez-Lopez, J.; Prieto-Conde, M.I.; Garcia-Alvarez, M.; et al. A Next-Generation Sequencing Strategy for Evaluating the Most Common Genetic Abnormalities in Multiple Myeloma. J. Mol. Diagn. 2017, 19, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Voigtlaender, M.; Janjetovic, S.; Thiele, B.; Alawi, M.; Marz, M.; Brandt, A.; Hansen, T.; Radloff, J.; Schon, G.; et al. Mutational landscape reflects the biological continuum of plasma cell dyscrasias. Blood Cancer J. 2017, 7, e537. [Google Scholar] [CrossRef] [PubMed]
- Moreaux, J.; Klein, B.; Bataille, R.; Descamps, G.; Maiga, S.; Hose, D.; Goldschmidt, H.; Jauch, A.; Reme, T.; Jourdan, M.; et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 2011, 96, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Tessoulin, B.; Eveillard, M.; Lok, A.; Chiron, D.; Moreau, P.; Amiot, M.; Moreau-Aubry, A.; le Gouill, S.; Pellat-Deceunynck, C. p53 dysregulation in B-cell malignancies: More than a single gene in the pathway to hell. Blood Rev. 2017, 31, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2017. [Google Scholar] [CrossRef] [PubMed]
- Surget, S.; Descamps, G.; Brosseau, C.; Normant, V.; Maiga, S.; Gomez-Bougie, P.; Gouy-Colin, N.; Godon, C.; Bene, M.C.; Moreau, P.; et al. RITA (Reactivating p53 and Inducing Tumor Apoptosis) is efficient against TP53abnormal myeloma cells independently of the p53 pathway. BMC Cancer 2014, 14, 437. [Google Scholar] [CrossRef] [PubMed]
- Tessoulin, B.; Descamps, G.; Moreau, P.; Maiga, S.; Lode, L.; Godon, C.; Marionneau-Lambot, S.; Oullier, T.; le Gouill, S.; Amiot, M.; et al. PRIMA-1Met induces myeloma cell death independent of p53 by impairing the GSH/ROS balance. Blood 2014, 124, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, B.; Millan, C.S.; Bonilla, F.; Dominguez, G. The TP73 complex network: Ready for clinical translation in cancer? Genes Chromosomes Cancer 2013, 52, 989–1006. [Google Scholar] [CrossRef] [PubMed]
- Zawacka-Pankau, J.; Kostecka, A.; Sznarkowska, A.; Hedstrom, E.; Kawiak, A. p73 tumor suppressor protein: A close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle 2010, 9, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Kawano, S.; Miller, C.W.; Gombart, A.F.; Bartram, C.R.; Matsuo, Y.; Asou, H.; Sakashita, A.; Said, J.; Tatsumi, E.; Koeffler, H.P. Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood 1999, 94, 1113–1120. [Google Scholar] [PubMed]
- Alexandrova, E.M.; Moll, U.M. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk. Lymphoma 2012, 53, 2116–2129. [Google Scholar] [CrossRef] [PubMed]
- Maas, A.M.; Bretz, A.C.; Mack, E.; Stiewe, T. Targeting p73 in cancer. Cancer Lett. 2013, 332, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Teoh, P.J.; Bi, C.; Sintosebastian, C.; Tay, L.S.; Fonseca, R.; Chng, W.J. PRIMA-1 targets the vulnerability of multiple myeloma of deregulated protein homeostasis through the perturbation of ER stress via p73 demethylation. Oncotarget 2016, 7, 61806–61819. [Google Scholar] [CrossRef] [PubMed]
- Surget, S.; Chiron, D.; Gomez-Bougie, P.; Descamps, G.; Menoret, E.; Bataille, R.; Moreau, P.; le Gouill, S.; Amiot, M.; Pellat-Deceunynck, C. Cell death via DR5, but not DR4, is regulated by p53 in myeloma cells. Cancer Res. 2012, 72, 4562–4573. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, D.; Meier, C.; Alla, V.; Putzer, B.M. A balancing act: Orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 2015, 34, 4287–4299. [Google Scholar] [CrossRef] [PubMed]
- Surget, S.; Lemieux-Blanchard, E.; Maiga, S.; Descamps, G.; le Gouill, S.; Moreau, P.; Amiot, M.; Pellat-Deceunynck, C. Bendamustine and melphalan kill myeloma cells similarly through reactive oxygen species production and activation of the p53 pathway and do not overcome resistance to each other. Leuk. Lymphoma 2014, 55, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Saha, M.N.; Jiang, H.; Yang, Y.; Reece, D.; Chang, H. PRIMA-1Met/APR-246 displays high antitumor activity in multiple myeloma by induction of p73 and Noxa. Mol. Cancer Ther. 2013, 12, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Moll, U.M.; Slade, N. p63 and p73: Roles in development and tumor formation. Mol. Cancer Res. 2004, 2, 371–386. [Google Scholar] [PubMed]
- Polager, S.; Ginsberg, D. p53 and E2f: Partners in life and death. Nat. Rev. Cancer 2009, 9, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Sayan, A.E.; Sreekumar, R.; Sayan, B.S. Regulation of p73 activity by post-translational modifications. Cell Death Dis. 2012, 3, e285. [Google Scholar] [CrossRef] [PubMed]
- Finzer, P.; Krueger, A.; Stohr, M.; Brenner, D.; Soto, U.; Kuntzen, C.; Krammer, P.H.; Rosl, F. HDAC inhibitors trigger apoptosis in HPV-positive cells by inducing the E2F-p73 pathway. Oncogene 2004, 23, 4807–4817. [Google Scholar] [CrossRef] [PubMed]



| HMCL | TP53 Status and Expression | TP73 Status and Expression | ||||
|---|---|---|---|---|---|---|
| Name | Translocation | TP53 | p53 | TP73 | Q-PCR | p73 |
| AMO1 | unknown | wt | + | wt | + | − |
| MDN | (11;14) | wt | + | wt | + | + |
| MM1S | (14;16) | wt | + | wt | + | + |
| NAN9 | (4;14) | wt | + | wt | − | − |
| NAN11 | (14;16) | wt | + | wt | + | + |
| XG6 | (16;22) | wt | + | wt | +/− | − |
| XG7 | (4;14) | wt | + | wt | +/− | nd |
| JJN3 | (14;16) | deletion | − | wt | + | + |
| KMS11 | (4;14) | deletion | − | wt | − | − |
| NAN8 | (4;14) | disrupted 1 | − | wt | − | − |
| JIM3 | (4;14) | R273C | + | wt | − | − |
| Karpas620 | (11;14) | C135Y | + | wt | − | nd |
| KMM1 | (6;14) | C135F | + | A211S | − | nd |
| KMS12PE | (11;14) | R337L | + | wt | − | − |
| LP1 | (4;14) | E286K | + | wt | − | − |
| L363 | (20;22) | S261T 2 | − | wt | − | − |
| NAN1 | (14;16) | E180STOP | − | wt | − | nd |
| NAN6 | (14;16) | Indel 3 | + 5 | wt | − | nd |
| NAN7 | (11;14) | Indel 4 | − | wt | − | nd |
| OPM2 | (4;14) | R175H | + | wt | − | − |
| SKMM2 | (11;14) | K132N | + | wt | − | nd |
| XG11 | (11;14) | C135Y | + | I626V | + | + |
| XG1 | (11;14) | Y126N | + | wt | +/− | nd |
| XG2 | unknown | C176Y | + | wt | +/− | nd |
| XG5 | (11;14) | R282W | + | wt | − | − |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillardin, P.-S.; Descamps, G.; Maiga, S.; Tessoulin, B.; Djamai, H.; Lucani, B.; Chiron, D.; Moreau, P.; Le Gouill, S.; Amiot, M.; et al. Decitabine and Melphalan Fail to Reactivate p73 in p53 Deficient Myeloma Cells. Int. J. Mol. Sci. 2018, 19, 40. https://doi.org/10.3390/ijms19010040
Gillardin P-S, Descamps G, Maiga S, Tessoulin B, Djamai H, Lucani B, Chiron D, Moreau P, Le Gouill S, Amiot M, et al. Decitabine and Melphalan Fail to Reactivate p73 in p53 Deficient Myeloma Cells. International Journal of Molecular Sciences. 2018; 19(1):40. https://doi.org/10.3390/ijms19010040
Chicago/Turabian StyleGillardin, Pierre-Samuel, Géraldine Descamps, Sophie Maiga, Benoit Tessoulin, Hanane Djamai, Benedetta Lucani, David Chiron, Philippe Moreau, Steven Le Gouill, Martine Amiot, and et al. 2018. "Decitabine and Melphalan Fail to Reactivate p73 in p53 Deficient Myeloma Cells" International Journal of Molecular Sciences 19, no. 1: 40. https://doi.org/10.3390/ijms19010040