Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor
Abstract
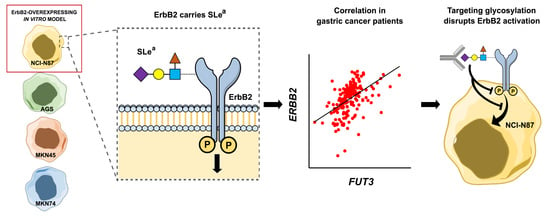
:1. Introduction
2. Results
2.1. Expression and Activation of the Human Epidermal Growth Factor Receptor Family in Gastric Cancer Cell Lines
2.2. Glycosyltransferase Expression in Gastric Cancer Cell Lines
2.3. Glycosylation Profile of ErbB2-Overexpressing NCI-N87 Cells
2.4. ErbB2 Glycan Signatures in NCI-N87 Cells
2.5. Interplay Between ErbB2 and Sialyl Lewis a (SLea) Expression
2.6. Inhibition of Glycosylation and Glycan Epitope-Blocking Disrupt ErbB2 Expression and Activation
3. Discussion
4. Materials and Methods
4.1. Gastric Cancer Cell Lines and Cell Culture
4.2. Western Blotting
4.3. Quantitative Real-Time PCR
4.4. Immunofluorescence Labeling of Glycan Epitopes
4.5. ErbB2 Immunoprecipitation
4.6. ErbB2 Identification by Matrix-Assisted Laser Desorption Ionization Time-Of-Flight (MALDI/TOF-TOF) Mass Spectrometry
4.7. ErbB2 Digestion with PNGase F
4.8. Tunicamycin and Cancer Antigen 19.9 (CA 19.9) MAb Treatment
4.9. Bioinformatic and Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BSA | Bovine serum albumin |
| CA 19.9 | Cancer antigen 19.9 |
| EGFR | Epidermal growth factor receptor |
| ErbB2 | Human epidermal growth factor receptor 2 |
| GC | Gastric cancer |
| GlcNAc | N-Acetylglucosamine |
| Lea | Lewis a |
| Leb | Lewis b |
| Lex | Lewis x |
| Ley | Lewis y |
| MAb | Monoclonal antibody |
| PBS | Phosphate buffered saline |
| RT-qPCR | Real-time quantitative PCR |
| RTKs | Receptor tyrosine kinases |
| SLea | Sialyl Lewis a |
| SLex | Sialyl Lewis x |
| STn | Sialyl Tn |
| TBS | Tris buffered saline |
| WB | Western blot |
References
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [PubMed]
- Waddell, T.; Verheij, M.; Allum, W.; Cunningham, D.; Cervantes, A.; Arnold, D. Gastric Cancer: Esmo–Esso–Estro Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2013, 24 (Suppl. 6), vi57–vi63. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Schwaederle, M.; Arguello, D.; Millis, S.Z.; Gatalica, Z.; Kurzrock, R. HER2 Expression Status in Diverse Cancers: Review of Results from 37,992 Patients. Cancer Metastasis Rev. 2015, 34, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in Gastric Cancer: A New Prognostic Factor and a Novel Therapeutic Target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.W.; Cho, H.S.; Eigenbrot, C.; Ferguson, K.M.; Garrett, T.P.; Leahy, D.J.; Lemmon, M.A.; Sliwkowski, M.X.; Ward, C.W.; Yokoyama, S. An Open-and-Shut Case? Recent Insights into the Activation of EGF/ErbB Receptors. Mol. Cell 2003, 12, 541–552. [Google Scholar] [CrossRef]
- Hynes, N.E.; Stern, D.F. The Biology of Erbb-2/Nue/Her-2 and Its Role in Cancer. Biochim. Biophys. Acta 1994, 1198, 165–184. [Google Scholar] [CrossRef]
- Hynes, N.E.; Lane, H.A. ERBB Receptors and Cancer: The Complexity of Targeted Inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T. Trastuzumab in Combination with Chemotherapy Versus Chemotherapy Alone for Treatment of Her2-Positive Advanced Gastric or Gastro-Oesophageal Junction Cancer (Toga): A Phase 3, Open-Label, Randomised Controlled Trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Roukos, D.H. Targeting Gastric Cancer with Trastuzumab: New Clinical Practice and Innovative Developments to Overcome Resistance. Ann. Surg. Oncol. 2010, 17, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Xu, R.H.; Bang, Y.J.; Hoff, P.M.; Liu, T.; Herráez-Baranda, L.A.; Xia, F.; Garg, A.; Shing, M.; Tabernero, J. Heloise: Phase Iiib Randomized Multicenter Study Comparing Standard-of-Care and Higher-Dose Trastuzumab Regimens Combined with Chemotherapy as First-Line Therapy in Patients with Human Epidermal Growth Factor Receptor 2-Positive Metastatic Gastric or Gastroesophageal Junction Adenocarcinoma. J. Clin. Oncol. 2017, 35, 2558–2567. [Google Scholar] [PubMed]
- Varki, A.; Stanley, P.; Schachter, H.; Taniguchi, N. Essentials of Glycobiology; Cold Spring Cold Spring Harbor Laboratory Press: Harbor, NY, USA, 2009. [Google Scholar]
- Pinho, S.S.; Reis, C.A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Carvalho, S.; Marcos-Pinto, R.; Magalhães, A.; Oliveira, C.; Gu, J.; Dinis-Ribeiro, M.; Carneiro, F.; Seruca, R.; Reis, C.A. Gastric Cancer: Adding Glycosylation to the Equation. Trends Mol. Med. 2013, 19, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Gomes, J.; Magalhães, A.; Reis, C.A. Glycomic Approaches for the Discovery of Targets in Gastrointestinal Cancer. Front. Oncol. 2016, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Contessa, J.N.; Bhojani, M.S.; Freeze, H.H.; Rehemtulla, A.; Lawrence, T.S. Inhibition of N-Linked Glycosylation Disrupts Receptor Tyrosine Kinase Signaling in Tumor Cells. Cancer Res. 2008, 68, 3803–3809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Yen, H.Y.; Chen, C.Y.; Chen, C.H.; Cheng, P.F.; Juan, Y.H.; Chen, C.H.; Khoo, K.H.; Yu, C.J.; Yang, P.C.; et al. Sialylation and Fucosylation of Epidermal Growth Factor Receptor Suppress Its Dimerization and Activation in Lung Cancer Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11332–11337. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Yokote, H.; Arao, T.; Maegawa, M.; Tanaka, K.; Fujita, Y.; Shimizu, C.; Hanafusa, T.; Fujiwara, Y.; Nishio, K. N-Glycan Fucosylation of Epidermal Growth Factor Receptor Modulates Receptor Activity and Sensitivity to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor. Cancer Sci. 2008, 99, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, J.; Ihara, H.; Miyoshi, E.; Honke, K.; Taniguchi, N. Core Fucosylation Regulates Epidermal Growth Factor Receptor-Mediated Intracellular Signaling. J. Biol. Chem. 2006, 281, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sambrooks, C.; Shrimal, S.; Khodier, C.; Flaherty, D.P.; Rinis, N.; Charest, J.C.; Gao, N.; Zhao, P.; Wells, L.; Lewis, T.A.; et al. Oligosaccharyltransferase Inhibition Induces Senescence in Rtk-Driven Tumor Cells. Nat. Chem. Biol. 2016, 12, 1023–1030. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Magalhães, A.; Adamczyk, B.; Jin, C.; Almeida, A.; Drici, L.; Ibáñez-Vea, M.; Gomes, C.; Ferreira, J.A.; Afonso, L.P.; et al. Glycomic Analysis of Gastric Carcinoma Cells Discloses Glycans as Modulators of Ron Receptor Tyrosine Kinase Activation in Cancer. Biochim. Biophys. Acta 2016, 1860, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Frei, A.P.; Jeon, O.Y.; Kilcher, S.; Moest, H.; Henning, L.M.; Jost, C.; Plückthun, A.; Mercer, J.; Aebersold, R.; Carreira, E.M.; et al. Direct Identification of Ligand-Receptor Interactions on Living Cells and Tissues. Nat. Biotechnol. 2012, 30, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Terasawa, K.; Kaneshiro, K.; Uchimura, H.; Yamamoto, R.; Fukuyama, Y.; Shimizu, K.; Sato, T.A.; Tanaka, K. Improvement of Mass Spectrometry Analysis of Glycoproteins by Maldi-Ms Using 3-Aminoquinoline/Α-Cyano-4-Hydroxycinnamic Acid. Anal. Bioanal. Chem. 2013, 405, 4289–4293. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.S.; Harduin-Lepers, A.; Magalhaes, A.; Machado, E.; Mendes, N.; Costa, L.T.; Matthiesen, R.; Almeida, R.; Costa, J.; Reis, C.A. Differential Expression of Α-2, 3-Sialyltransferases and Α-1, 3/4-Fucosyltransferases Regulates the Levels of Sialyl Lewis a and Sialyl Lewis X in Gastrointestinal Carcinoma Cells. Int. J. Biochem. Cell Biol. 2010, 42, 80–89. [Google Scholar] [PubMed]
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Narimatsu, H. Fucosyltransferase 3. Gdp-Fucose Lactosamine Α1, 3/4-Fucosyltransferase. Lea and Leb Histo-Blood Groups (Fut3, Lewis Enzyme). In Handbook of Glycosyltransferases and Related Genes; Springer Science & Business Media: Berlin, Germany, 2014; pp. 531–539. [Google Scholar]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Klinger, M.; Farhan, H.; Just, H.; Drobny, H.; Himmler, G.; Loibner, H.; Mudde, G.C.; Freissmuth, M.; Sexl, V. Antibodies Directed against Lewis-Y Antigen Inhibit Signaling of Lewis-Y Modified Erbb Receptors. Cancer Res. 2004, 64, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Tanner, M.; Hollmén, M.; Junttila, T.T.; Kapanen, A.I.; Tommola, S.; Soini, Y.; Helin, H.; Salo, J.; Joensuu, H.; Sihvo, E.; et al. Amplification of HER-2 in Gastric Carcinoma: Association with Topoisomerase IIα Gene Amplification, Intestinal Type, Poor Prognosis and Sensitivity to Trastuzumab. Ann. Oncol. 2005, 16, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.J.; Kumagai, T.; Berezov, A.; Murali, R.; Greene, M.I. HER2/Neu: Mechanisms of Dimerization/Oligomerization. Oncogene 2000, 19, 6093–6101. [Google Scholar] [CrossRef] [PubMed]
- Graus-Porta, D.; Beerli, R.R.; Hynes, N.E. Single-Chain Antibody-Mediated Intracellular Retention of Erbb-2 Impairs Neu Differentiation Factor and Epidermal Growth Factor Signaling. Mol. Cell. Biol. 1995, 15, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Tzahar, E.; Waterman, H.; Chen, X.; Levkowitz, G.; Karunagaran, D.; Lavi, S.; Ratzkin, B.J.; Yarden, Y. A Hierarchical Network of Interreceptor Interactions Determines Signal Transduction by Neu Differentiation Factor/Neuregulin and Epidermal Growth Factor. Mol. Cell. Biol. 1996, 16, 5276–5287. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, T.; Ikeda, Y.; Taniguchi, N. The Asn-420-Linked Sugar Chain in Human Epidermal Growth Factor Receptor Suppresses Ligand-Independent Spontaneous Oligomerization Possible Role of a Specific Sugar Chain in Controllable Receptor Activation. J. Biol. Chem. 2000, 275, 21988–21994. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.; Cohen, S.; Bishayee, S. Glycosylation-Induced Conformational Modification Positively Regulates Receptor-Receptor Association a Study with an Aberrant Epidermal Growth Factor Receptor (Egfrviii/Δegfr) Expressed in Cancer Cells. J. Biol. Chem. 2001, 276, 5375–5383. [Google Scholar] [CrossRef] [PubMed]
- Yokoe, S.; Takahashi, M.; Asahi, M.; Lee, S.H.; Li, W.; Osumi, D.; Miyoshi, E.; Taniguchi, N. The Asn418-Linked N-Glycan of Erbb3 Plays a Crucial Role in Preventing Spontaneous Heterodimerization and Tumor Promotion. Cancer Res. 2007, 67, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yokoe, S.; Asahi, M.; Lee, S.H.; Li, W.; Osumi, D.; Miyoshi, E.; Taniguchi, N. N-Glycan of Erbb Family Plays a Crucial Role in Dimer Formation and Tumor Promotion. Biochim. Biophys. Acta 2008, 1780, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Ashkani, J.; Naidoo, K.J. Glycosyltransferase Gene Expression Profiles Classify Cancer Types and Propose Prognostic Subtypes. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.; Rossez, Y.; Robbe-Masselot, C.; Maes, E.; Gomes, J.; Shevtsova, A.; Bugaytsova, J.; Borén, T.; Reis, C.A. Muc5ac Gastric Mucin Glycosylation Is Shaped by Fut2 Activity and Functionally Impacts Helicobacter Pylori Binding. Sci. Rep. 2016, 6, 25575. [Google Scholar] [CrossRef] [PubMed]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis X in Gastrointestinal Cancers. Biology 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, W.; Fregien, N.; Pierce, M. The Her-2/Neu Oncogene Stimulates the Transcription of N-Acetylglucosaminyltransferase V and Expression of Its Cell Surface Oligosaccharide Products. Oncogene 1998, 17, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Buckhaults, P.; Chen, L.; Fregien, N.; Pierce, M. Transcriptional Regulation of N-acetylglucosaminyltransferase V by the Srconcogene. J. Biol. Chem. 1997, 272, 19575–19581. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.B.; Johnson, H.; Randolph, M.; Nagy, T.; Blalock, R.; Pierce, M. Specific Posttranslational Modification Regulates Early Events in Mammary Carcinoma Formation. Proc. Natl Acad. Sci. USA 2010, 107, 21116–21121. [Google Scholar] [CrossRef] [PubMed]
- Hadari, Y.R.; Doody, J.F.; Wang, Y.; Patel, S.N.; Apblett, R.L.; Loizos, N.; Pereira, D.S.; Witte, L.; Bohlen, P.; Hicklin, D.J. The Igg1 Monoclonal Antibody Cetuximab Induces Degradation of the Epidermal Growth Factor Receptor. In Proceedings of the ASCO Gastrointestinal Cancers Symposium, San Francisco, CA, USA, 22–24 January 2004. [Google Scholar]
- Anderson, W.F.; Camargo, M.C.; Fraumeni, J.F., Jr.; Correa, P.; Rosenberg, P.S.; Rabkin, C.S. Age-Specific Trends in Incidence of Noncardia Gastric Cancer in US Adults. JAMA 2010, 303, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Weston, B.W.; Hiller, K.M.; Mayben, J.P.; Manousos, G.A.; Bendt, K.M.; Liu, R.; Cusack, J.C., Jr. Expression of Human Α (1, 3) Fucosyltransferase Antisense Sequences Inhibits Selectin-Mediated Adhesion and Liver Metastasis of Colon Carcinoma Cells. Cancer Res. 1999, 59, 2127–2135. [Google Scholar] [PubMed]
- Opolski, A.; Laskowska, A.; Madej, J.; Wietrzyk, J.; Kłopocki, A.; Radzikowski, C.; Ugorski, M. Metastatic Potential of Human Cx-1 Colon Adenocarcinoma Cells is Dependent on the Expression of Sialosyl Le a Antigen. Clin. Exp. Metastasis 1998, 16, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, J.; Sondén, B.; Hurtig, M.; Olfat, F.O.; Forsberg, L.; Roche, N.; Angstrom, J.; Larsson, T.; Teneberg, S.; Karlsson, K.A.; et al. Helicobacter Pylori Saba Adhesin in Persistent Infection and Chronic Inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Hiller, K.M.; Mayben, J.P.; Bendt, K.M.; Manousos, G.A.; Senger, K.; Cameron, H.S.; Weston, B.W. Transfection of Α (1, 3) Fucosyltransferase Antisense Sequences Impairs the Proliferative and Tumorigenic Ability of Human Colon Carcinoma Cells. Mol. Carcinog. 2000, 27, 280–288. [Google Scholar] [CrossRef]
- Terraneo, L.; Avagliano, L.; Caretti, A.; Bianciardi, P.; Tosi, D.; Bulfamante, G.P.; Samaja, M.; Trinchera, M. Expression of Carbohydrate-Antigen Sialyl-Lewis a on Colon Cancer Cells Promotes Xenograft Growth and Angiogenesis in Nude Mice. Int. J. Biochem. Cell Biol. 2013, 45, 2796–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinberg, W. The Clinical Utility of the Ca 19–9 Tumor-Associated Antigen. Am. J. Gastroenterol. 1990, 85, 350–355. [Google Scholar] [PubMed]
- Holdenrieder, S.; Pagliaro, L.; Morgenstern, D.; Dayyani, F. Clinically Meaningful Use of Blood Tumor Markers in Oncology. BioMed Res. Int. 2016, 2016, 9795269. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in Glycosylation as Biomarkers for Cancer Detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Perng, C.L.; Lin, H.J.; Lee, S.D. Serum C-Erbb-2 Oncoprotein in the Diagnosis of Gastric Cancer in Comparison with Ca 19–9, Cea, Tpa, Ca 125 and Afp. Zhonghua yi xue za zhi 1994, 54, 82–86. [Google Scholar] [PubMed]
- Molina, R.; Jo, J.; Filella, X.; Bruix, J.; Castells, A.; Hague, M.; Ballesta, A.M. Serum Levels of C-Erbb-2 (Her-2/Neu) in Patients with Malignant and Non-Malignant Diseases. Tumor Biol. 1997, 18, 188–196. [Google Scholar] [CrossRef]
- Kono, K.; Naganuma, H.; Sekikawa, T.; Amemiya, H.; Takahashi, A.; Iizuka, H.; Matsumoto, Y. Serum Level of Her-2/Neu in Patients with Gastric Cancer: Correlation with Her-2/Neu Overexpression in Gastric Carcinoma Tissue. Tumor Biol. 2000, 21, 139–144. [Google Scholar] [CrossRef]
- Wong, H.; Yau, T. Molecular Targeted Therapies in Advanced Gastric Cancer: Does Tumor Histology Matter? Therap. Adv. Gastroenterol. 2013, 6, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Colcher, D.; Hand, P.H.; Nuti, M.; Schlom, J. A Spectrum of Monoclonal Antibodies Reactive with Human Mammary Tumor Cells. Proc. Natl. Acad. Sci. USA 1981, 78, 3199–3203. [Google Scholar] [CrossRef] [PubMed]
- Young, W.W., Jr.; Portoukalian, J.; Hakomori, S. Two Monoclonal Anticarbohydrate Antibodies Directed to Glycosphingolipids with a Lacto-N-Glycosyl Type II Chain. J. Biol. Chem. 1981, 256, 10967–10972. [Google Scholar] [PubMed]
- Fukushi, Y.; Hakomori, S.; Nudelman, E.; Cochran, N. Novel Fucolipids Accumulating in Human Adenocarcinoma. II. Selective Isolation of Hybridoma Antibodies That Differentially Recognize Mono-, Di-, and Trifucosylated Type 2 Chain. J. Biol. Chem. 1984, 259, 4681–4685. [Google Scholar] [PubMed]
- Abe, K.; McKibbin, J.M.; Hakomori, S. The Monoclonal Antibody Directed to Difucosylated Type 2 Chain (Fuc Alpha 1 Leads to 2gal Beta 1 Leads to 4 [Fuc Alpha 1 Leads to 3] Glcnac; Y Determinant). J. Biol. Chem. 1983, 258, 11793–11797. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2-Δδct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ferreirinha, P.; Correia, A.; Teixeira-Coelho, M.; Osório, H.; Teixeira, L.; Rocha, A.; Vilanova, M. Mucosal Immunization Confers Long-Term Protection against Intragastrically Established Neospora Caninum Infection. Vaccine 2016, 34, 6250–6258. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. Oncomine: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Ooi, W.F.; Xing, M.; Xu, C.; Yao, X.; Ramlee, M.K.; Lim, M.C.; Cao, F.; Lim, K.; Babu, D.; Poon, L.F. Epigenomic Profiling of Primary Gastric Adenocarcinoma Reveals Super-Enhancer Heterogeneity. Nat. Commun. 2016, 7, 12983. [Google Scholar] [CrossRef] [PubMed]






| Antibody Clone/Lectin | Antigen | Working Dilution | Supplier | |
|---|---|---|---|---|
| IF | WB | |||
| 29D8 | ErbB2 | - | 1:1000 | Cell Signaling Technology |
| Polyclonal | pErbB2 (Tyr1221/1222) | - | 1:1000 | Cell Signaling Technology |
| D38B1 | EGFR (ErbB1) | - | 1:1000 | Cell Signaling Technology |
| D7A5 | pEGFR (Tyr1068) | - | 1:1000 | Cell Signaling Technology |
| D22C5 | ErbB3 | - | 1:2000 | Cell Signaling Technology |
| 111B2 | ErbB4 | - | 1:1000 | Cell Signaling Technology |
| DM1A | α-Tubulin | - | 1:10,000 | Sigma-Aldrich |
| CA 19.9 (241) | SLea | 1:500 | 1:1000 | Santa Cruz Biotechnology |
| SPM279 | Lea | - | 1:200 | Santa Cruz Biotechnology |
| CSLEX1 | SLex | 1:80 | 1:1000 | BD PharmingenTM |
| B72.3 | STn | 1:5 | - | [56] |
| CA3F4 | Lea | 1:5 | - | [57] |
| BG6 (T218) | Leb | 1:50 | 1:200 | Signet |
| SH1 | Lex | 1:5 | Undiluted | [58] |
| AH6 | Ley | 1:2 | Undiluted | [59] |
| Aleuria aurantia lectin (AAL) | Fucα6GlcNAc | 1:500 | 1:3000 | Vector Labs |
| Phaseolus vulgaris leucoagglutinin (PHA-L) | Galβ4GlcNAcβ6 (GlcNAcβ2Manα3) Manα3 | 1:500 | 1:2000 | Vector Labs |
| Phaseolus vulgaris erythroagglutinin (PHA-E) | Galβ4GlcNAcβ2Manα6 (GlcNAcβ4) (GlcNAcβ4Manα3) Manβ4 | 1:500 | 1:2000 | Vector Labs |
| Concanavalin A (Con A) | αMan, αGlc | 1:500 | 1:2000 | Vector Labs |
| Sambucus nigra lectin (SNA) | Neu5Acα6Gal/GalNAc | 1:500 | 1:3000 | Vector Labs |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, H.O.; Balmaña, M.; Mereiter, S.; Osório, H.; Gomes, J.; Reis, C.A. Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor. Int. J. Mol. Sci. 2017, 18, 2262. https://doi.org/10.3390/ijms18112262
Duarte HO, Balmaña M, Mereiter S, Osório H, Gomes J, Reis CA. Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor. International Journal of Molecular Sciences. 2017; 18(11):2262. https://doi.org/10.3390/ijms18112262
Chicago/Turabian StyleDuarte, Henrique O., Meritxell Balmaña, Stefan Mereiter, Hugo Osório, Joana Gomes, and Celso A. Reis. 2017. "Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor" International Journal of Molecular Sciences 18, no. 11: 2262. https://doi.org/10.3390/ijms18112262