Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation
Abstract
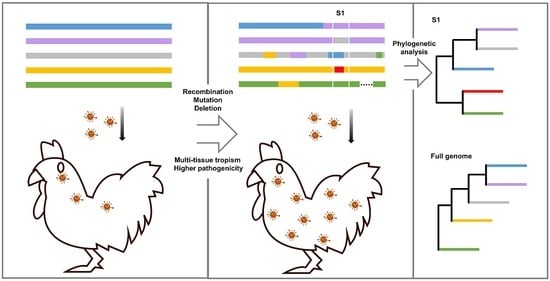
:1. Introduction
2. IBV Classification
3. Evolution Mechanism of IBV
4. Asia
5. Africa
6. Europe
7. America
8. Oceania
9. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schalk, A.; Hawn, M. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931, 78, 413–422. [Google Scholar]
- Cavanagh, D.; Davis, P.J.; Mockett, A.P.A. Amino-Acids within Hypervariable Region-1 of Avian Coronavirus Ibv (Massachusetts Serotype) Spike Glycoprotein Are Associated with Neutralization Epitopes. Virus Res. 1988, 11, 141–150. [Google Scholar] [CrossRef]
- Cavanagh, D.; Elus, M.M.; Cook, J.K. Relationship between sequence variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection in vivo. Avian Pathol. 1997, 26, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D.; Davis, P.J. Coronavirus IBV: Removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986, 67, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Promkuntod, N.; Van Eijndhoven, R.; De Vrieze, G.; Gröne, A.; Verheije, M. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology 2014, 448, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Casais, R.; Dove, B.; Cavanagh, D.; Britton, P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003, 77, 9084–9089. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cai, H.; Pan, J.A.; Xiang, N.; Tien, P.; Ahola, T.; Guo, D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA 2009, 106, 3484–3489. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Chen, G.; Guo, B.; Cheng, G.; Tang, H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008, 18, 1105–1113. [Google Scholar] [PubMed]
- Seo, S.H.; Wang, L.; Smith, R.; Collisson, E.W. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. J. Virol. 1997, 71, 7889–7894. [Google Scholar] [PubMed]
- Yang, T.; Wang, H.-N.; Wang, X.; Tang, J.-N.; Lu, D.; Zhang, Y.-F.; Guo, Z.-C.; Li, Y.-L.; Gao, R.; Kang, R.-M. The protective immune response against infectious bronchitis virus induced by multi-epitope based peptide vaccines. Biosci. Biotechnol. Biochem. 2009, 73, 1500–1504. [Google Scholar] [CrossRef] [PubMed]
- Archetti, I.; Horsfall, F.L., Jr. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950, 92, 441. [Google Scholar] [CrossRef] [PubMed]
- Ruano, M.; El-Attrache, J.; Villegas, P. A rapid-plate hemagglutination assay for the detection of infectious bronchitis virus. Avian Dis. 2000, 44, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Karaca, K.; Naqi, S. A monoclonal antibody blocking ELISA to detect serotype-specific infectious bronchitis virus antibodies. Vet. Microbiol. 1993, 34, 249–257. [Google Scholar] [CrossRef]
- Chen, H.W.; Wang, C.H.; Cheng, I.C. A type-specific blocking ELISA for the detection of infectious bronchitis virus antibody. J. Virol. Methods 2011, 173, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed]
- Ladman, B.S.; Loupos, A.B.; Gelb, J., Jr. Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization. Avian Pathol. 2006, 35, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D.; Davis, P.; Cook, J.K.; Li, D.; Kant, A.; Koch, G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992, 21, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016, 39, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D.; Naqi, S. Infectious Bronchitis; John Wiley & Sons: Hoboken, NJ, USA, 2003; Volume 11, pp. 101–119. [Google Scholar]
- Chen, Y.; Jiang, L.; Zhao, W.; Liu, L.; Zhao, Y.; Shao, Y.; Li, H.; Han, Z.; Liu, S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017, 198, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, M.; Tian, X.; Shao, H.; Qian, K.; Ye, J.; Qin, A. Identification of a novel recombinant virulent avian infectious bronchitis virus. Vet. Microbiol. 2017, 199, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, X.; Xu, P.; Zhou, L.; Zhang, Z.; Wang, H. Genome sequence and origin analyses of the recombinant novel IBV virulent isolate SAIBK2. Virus Genes 2016, 52, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, T.; Han, Z.; Liang, S.; Xu, Y.; Xu, Q.; Chen, Y.; Zhao, Y.; Shao, Y.; Li, H.; et al. Molecular and antigenic characteristics of Massachusetts genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2015, 181, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Hu, Y.X.; Jin, J.H.; Zhao, Y.; Zhao, J.; Zhang, G.Z. Pathogenicity of virulent infectious bronchitis virus isolate YN on hen ovary and oviduct. Vet. Microbiol. 2016, 193, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Hu, Y.; Ma, Z.; Yu, Q.; Zhao, J.; Liu, X.; Zhang, G. Virulent avian infectious bronchitis virus, People’s Republic of China. Emerg. Infect. Dis. 2012, 18, 1994–2001. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Liu, X.Y.; Zhao, Y.; Chen, Y.; Zhao, J.; Zhang, G.Z. Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virol. J. 2016, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.H.; Chen, Y.; Zhao, J.; Xu, G.; Zhao, Y.; Zhang, G.Z. Pathogenicity of a TW-Like Strain of Infectious Bronchitis Virus and Evaluation of the Protection Induced against It by a QX-Like Strain. Front. Microbiol. 2016, 7, 1653. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, Q.; Zhao, W.; Chen, Y.; Zhang, T.; Han, Z.; Xu, Q.; Kong, X.; Liu, S. Serotype, antigenicity, and pathogenicity of a naturally recombinant TW I genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2016, 191, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhang, T.; Xu, Q.; Gao, M.; Chen, Y.; Wang, Q.; Zhao, Y.; Shao, Y.; Li, H.; Kong, X.; et al. Altered pathogenicity of a tl/CH/LDT3/03 genotype infectious bronchitis coronavirus due to natural recombination in the 5′- 17kb region of the genome. Virus Res. 2016, 213, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, Q.; Han, Z.; Liu, X.; Li, H.; Guo, H.; Sun, N.; Shao, Y.; Kong, X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect. Genet. Evol. 2014, 23, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Tang, M.; Jiang, Y.; Chen, X.; Shen, X.; Li, J.; Dai, Y.; Zou, J. Complete genome sequence of a novel infectious bronchitis virus strain circulating in China with a distinct S gene. Virus Genes 2014, 49, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zou, N.; Wang, F.; Guo, M.; Liu, P.; Wen, X.; Cao, S.; Huang, Y. Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a, ORF 5b, and nucleocapsid protein gene sequences. Virus Genes 2013, 46, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.T.; Ma, B.C. Complete genome sequence of a nephropathogenic infectious bronchitis virus strain isolated in china. Genome Announc. 2013, 1, e00815-13. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Li, Y.T.; Chen, Y.T.; Chen, T.C.; Hu, C.J.; Chen, H.W. Identification of an infectious bronchitis coronavirus strain exhibiting a classical genotype but altered antigenicity, pathogenicity, and innate immunity profile. Sci. Rep. 2016, 6, 37725. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, Y.; Horiuchi, T.; Harada, M.; Tanimura, C.; Seki, Y.; Kuroda, Y.; Yagyu, K.; Nakamura, S.; Suzuki, S. Isolation of 4/91 type of infectious bronchitis virus as a new variant in Japan and efficacy of vaccination against 4/91 type field isolate. Avian Dis. 2008, 52, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Kwon, H.J.; Kim, I.H.; Mo, M.L.; Kim, J.H. Comparative genomics of Korean infectious bronchitis viruses (IBVs) and an animal model to evaluate pathogenicity of IBVs to the reproductive organs. Viruses 2012, 4, 2670–2683. [Google Scholar] [CrossRef] [PubMed]
- Khataby, K.; Kichou, F.; Loutfi, C.; Ennaji, M.M. Assessment of pathogenicity and tissue distribution of infectious bronchitis virus strains (Italy 02 genotype) isolated from moroccan broiler chickens. BMC Vet. Res. 2016, 12, 94. [Google Scholar] [CrossRef] [PubMed]
- Zanaty, A.; Arafa, A.S.; Hagag, N.; El-Kady, M. Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J. Virol. 2016, 5, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Zanaty, A.; Naguib, M.M.; El-Husseiny, M.H.; Mady, W.; Hagag, N.; Arafa, A.S. The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra-genotypic recombination. Arch. Virol. 2016, 161, 3583–3587. [Google Scholar] [CrossRef] [PubMed]
- Franca, M.; Woolcock, P.R.; Yu, M.; Jackwood, M.W.; Shivaprasad, H.L. Nephritis associated with infectious bronchitis virus Cal99 variant in game chickens. Avian Dis. 2011, 55, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Chacon, J.L.; Assayag, M.S., Jr.; Revolledo, L.; Astolfi-Ferreira, C.S.; Vejarano, M.P.; Jones, R.C.; Piantino Ferreira, A.J. Pathogenicity and molecular characteristics of infectious bronchitis virus (IBV) strains isolated from broilers showing diarrhoea and respiratory disease. Brit. Poult. Sci. 2014, 55, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Bonci, M.; Bano, L.; Bano, L.; Valastro, V.; Vascellari, M.; Capua, I.; Terregino, C. Diagnostic and clinical observation on the infectious bronchitis virus strain Q1 in Italy. Vet. Ital. 2013, 49, 347–355. [Google Scholar] [PubMed]
- Franzo, G.; Listorti, V.; Naylor, C.J.; Lupini, C.; Laconi, A.; Felice, V.; Drigo, M.; Catelli, E.; Cecchinato, M. Molecular investigation of a full-length genome of a Q1-like IBV strain isolated in Italy in 2013. Virus Res. 2015, 210, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, K.; Wilkins, M.; Forrester, A.; Lemiere, S.; Cserep, T.; McMullin, P.; Jones, R.C. QX-like infectious bronchitis virus isolated from cases of proventriculitis in commercial broilers in England. Vet. Rec. 2012, 171, 597. [Google Scholar] [CrossRef] [PubMed]
- Hewson, K.A.; Noormohammadi, A.H.; Devlin, J.M.; Browning, G.F.; Schultz, B.K.; Ignjatovic, J. Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathol. 2014, 43, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Quinteros, J.A.; Lee, S.W.; Markham, P.F.; Noormohammadi, A.H.; Hartley, C.A.; Legione, A.R.; Coppo, M.J.; Vaz, P.K.; Browning, G.F. Full genome analysis of Australian infectious bronchitis viruses suggests frequent recombination events between vaccine strains and multiple phylogenetically distant avian coronaviruses of unknown origin. Vet. Microbiol. 2016, 197, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Mayr, E. Toward a New Philosophy of Biology: Observations of an Evolutionist; Harvard University Press: Cambridge, MA, USA, 1988. [Google Scholar]
- Smith, E.C.; Blanc, H.; Vignuzzi, M.; Denison, M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: Evidence for proofreading and potential therapeutics. PLoS Pathog. 2013, 9, e1003565. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [PubMed]
- Toro, H.; van Santen, V.L.; Jackwood, M.W. Genetic diversity and selection regulates evolution of infectious bronchitis virus. Avian Dis. 2012, 56, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, R.A.; Van Santen, V.L.; Toro, H. Host intraspatial selection of infectious bronchitis virus populations. Avian Dis. 2010, 54, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Shahwan, K.; Hesse, M.; Mork, A.K.; Herrler, G.; Winter, C. Sialic acid binding properties of soluble coronavirus spike (S1) proteins: Differences between infectious bronchitis virus and transmissible gastroenteritis virus. Viruses 2013, 5, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, H.; Zhao, J.; Zhong, Q.; Jin, J.H.; Zhang, G.Z. Evolution of infectious bronchitis virus in China over the past two decades. J. Gen. Virol. 2016, 97, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Tsai, C.T. Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Arch. Virol. 1996, 141, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Huang, Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000, 145, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S.M.; Wang, C.H.; Hou, M.H.; Huang, Y.P.; Kao, H.W.; Su, H.L. Evolution of infectious bronchitis virus in Taiwan: Characterisation of RNA recombination in the nucleocapsid gene. Vet. Microbiol. 2010, 144, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-W.; Huang, Y.-P.; Wang, C.-H. Identification of Taiwan and China-like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res. 2009, 140, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; Tsai, H.F.; Wang, C.H. Detection of infectious bronchitis virus strains similar to Japan in Taiwan. J. Vet. Med. Sci. 2016, 78, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Huang, Y.P.; Wang, C.H. Identification of intertypic recombinant infectious bronchitis viruses from slaughtered chickens. Poult. Sci. 2010, 89, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Shao, Y.; Sun, C.; Han, Z.; Liu, X.; Guo, H.; Liu, X.; Kong, X.; Liu, S. Genetic diversity of avian infectious bronchitis coronavirus in recent years in China. Avian Dis. 2012, 56, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Yudong, W.; YongLin, W.; Zichun, Z.; GenChe, F.; Yihai, J.; Xiange, L.; Jiang, D.; Shushuang, W. Isolation and identification of glandular stomach type IBV (QX IBV) in chickens. Chin. J. Anim. Quarantine 1998, 15, 1–3. [Google Scholar]
- Sun, C.; Han, Z.; Ma, H.; Zhang, Q.; Yan, B.; Shao, Y.; Xu, J.; Kong, X.; Liu, S. Phylogenetic analysis of infectious bronchitis coronaviruses newly isolated in China, and pathogenicity and evaluation of protection induced by Massachusetts serotype H120 vaccine against QX-like strains. Avian Pathol. 2011, 40, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Sun, C.; Yan, B.; Zhang, X.; Wang, Y.; Li, C.; Zhang, Q.; Ma, Y.; Shao, Y.; Liu, Q.; et al. A 15-year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011, 11, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Xue, Y.; Wang, F.; Chen, F.; Shu, D.; Xie, Q. Analysis of S1 gene of avian infectious bronchitis virus isolated in southern China during 2011–2012. Virus Genes 2014, 49, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xue, C.; Chen, F.; Qin, J.; Xie, Q.; Bi, Y.; Cao, Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004–2008. Vet. Microbiol. 2010, 143, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Xie, J.; Chen, F.; Shu, D.; Zuo, K.; Xue, C.; Qin, J.; Li, H.; Bi, Y.; Ma, J. Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008–2009. Virol. J. 2011, 8, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, Y.; Wang, H.; Zeng, F.; Yang, X.; Zhang, Y.; Zhang, A. Molecular detection and Smoothing spline clustering of the IBV strains detected in China during 2011–2012. Virus Res. 2016, 211, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.L.; Li, M.; Huang, B.C.; Fan, W.S.; Wei, P.; Wei, T.C.; Cheng, Q.Y.; Wei, Z.J.; Lang, Y.H. Molecular characterization of major structural protein genes of avian coronavirus infectious bronchitis virus isolates in southern china. Viruses 2013, 5, 3007–3020. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, X.; Yao, K.C.; Du, L.J.; Liu, P.; Yan, Q.G.; Wen, Y.P.; Cao, S.J.; Han, X.F.; Huang, Y. Phylogenetic and antigenic analysis of avian infectious bronchitis virus in southwestern China, 2012–2016. Infect. Genet. Evol. 2016, 45, 11–19. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Li, M.; Wei, P.; Mo, M.L.; Wei, T.C.; Li, K.R. Complete genome sequence of an infectious bronchitis virus chimera between cocirculating heterotypic strains. J. Virol. 2012, 86, 13887–13888. [Google Scholar] [CrossRef] [PubMed]
- Zou, N.L.; Zhao, F.F.; Wang, Y.P.; Liu, P.; Cao, S.J.; Wen, X.T.; Huang, Y. Genetic analysis revealed LX4 genotype strains of avian infectious bronchitis virus became predominant in recent years in Sichuan area, China. Virus Genes 2010, 41, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Qin, J.; Chen, F.; Xie, Q.; Bi, Y.; Cao, Y.; Xue, C. Phylogenetic analysis of the S1 glycoprotein gene of infectious bronchitis viruses isolated in China during 2009–2010. Virus Genes 2012, 44, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Yamaguchi, S.; Imada, T. Existence of avian infectious bronchitis virus with a European-prevalent 4/91 genotype in Japan. J. Vet. Med. Sci. 2008, 70, 1341–1344. [Google Scholar]
- Shimazaki, Y.; Watanabe, Y.; Harada, M.; Seki, Y.; Kuroda, Y.; Fukuda, M.; Honda, E.; Suzuki, S.; Nakamura, S. Genetic analysis of the S1 gene of 4/91 type infectious bronchitis virus isolated in Japan. J. Vet. Med. Sci. 2009, 71, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Mase, M.; Kawanishi, N.; Ootani, Y.; Murayama, K.; Karino, A.; Inoue, T.; Kawakami, J. A novel genotype of avian infectious bronchitis virus isolated in Japan in 2009. J. Vet. Med. Sci. 2010, 72, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.-H.; Lee, H.-J.; Lee, D.-H.; Lee, Y.-N.; Park, J.-K.; Youn, H.-N.; Kim, M.-S.; Lee, J.-B.; Park, S.-Y.; Choi, I.-S. An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect. Genet. Evol. 2011, 11, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Kwon, H.J.; Choi, K.S.; Kim, J.H. Comparative genomics of QX-like infectious bronchitis viruses in Korea. Arch. Virol. 2017, 162, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Pohuang, T.; Chansiripornchai, N.; Tawatsin, A.; Sasipreeyajan, J. Sequence analysis of S1 genes of infectious bronchitis virus isolated in Thailand during 2008–2009: Identification of natural recombination in the field isolates. Virus Genes 2011, 43, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Promkuntod, N.; Thongmee, S.; Yoidam, S. Analysis of the S1 gene of the avian infectious bronchitis virus (IBV) reveals changes in the IBV genetic groups circulating in southern Thailand. Res. Vet. Sci. 2015, 100, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.H.; Bhimani, M.P.; Bhanderi, B.B.; Jhala, M.K. Isolation and molecular characterization of nephropathic infectious bronchitis virus isolates of Gujarat state, India. Virus Disease 2015, 26, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Sumi, V.; Singh, S.D.; Dhama, K.; Gowthaman, V.; Barathidasan, R.; Sukumar, K. Isolation and molecular characterization of infectious bronchitis virus from recent outbreaks in broiler flocks reveals emergence of novel strain in India. Trop. Anim. Health Prod. 2012, 44, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, K.; Ball, C.; Forrester, A. Genotypes of infectious bronchitis viruses circulating in the Middle East between 2009 and 2014. Virus Res. 2015, 210, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Seger, W.; GhalyanchiLangeroudi, A.; Karimi, V.; Madadgar, O.; Marandi, M.V.; Hashemzadeh, M. Genotyping of infectious bronchitis viruses from broiler farms in Iraq during 2014–2015. Arch. Virol. 2016, 161, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Ababneh, M.; Dalab, A.E.; Alsaad, S.; Al-Zghoul, M. Presence of Infectious Bronchitis Virus Strain CK/CH/LDL/97I in the Middle East. ISRN Vet. Sci. 2012, 2012, 201721. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Monne, I.; Terregino, C.; Cattoli, G.; Hodobo, C.T.; Gadaga, B.; Makaya, P.V.; Mdlongwa, E.; Swiswa, S. QX-like infectious bronchitis virus in Africa. Vet. Rec. 2011, 169, 589. [Google Scholar] [CrossRef] [PubMed]
- Fellahi, S.; Ducatez, M.; El Harrak, M.; Guerin, J.L.; Touil, N.; Sebbar, G.; Bouaiti el, A.; Khataby, K.; Ennaji, M.M.; El-Houadfi, M. Prevalence and molecular characterization of avian infectious bronchitis virus in poultry flocks in Morocco from 2010 to 2014 and first detection of Italy 02 in Africa. Avian Pathol. 2015, 44, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Abro, S.H.; Renstrom, L.H.; Ullman, K.; Belak, S.; Baule, C. Characterization and analysis of the full-length genome of a strain of the European QX-like genotype of infectious bronchitis virus. Arch. Virol. 2012, 157, 1211–1215. [Google Scholar] [CrossRef] [PubMed]
- Irvine, R.M.; Cox, W.J.; Ceeraz, V.; Reid, S.M.; Ellis, R.J.; Jones, R.M.; Errington, J.; Wood, A.M.; McVicar, C.; Clark, M.I. Detection of IBV QX in commercial broiler flocks in the UK. Vet. Rec. 2010, 167, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Pohjola, L.K.; Ek-Kommonen, S.C.; Tammiranta, N.E.; Kaukonen, E.S.; Rossow, L.M.; Huovilainen, T.A. Emergence of avian infectious bronchitis in a non-vaccinating country. Avian Pathol. 2014, 43, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Krapez, U.; Slavec, B.; Barlic-Maganja, D.; Rojs, O.Z. Molecular analysis of infectious bronchitis viruses isolated in Slovenia between 1990 and 2005: A retrospective study. Virus Genes 2010, 41, 414–416. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, E.V.; Bochkov, Y.A.; Shcherbakova, L.O.; Nikonova, Z.B.; Zinyakov, N.G.; Elatkin, N.P.; Mudrak, N.S.; Borisov, A.V.; Drygin, V.V. Molecular characterization of infectious bronchitis virus isolates from Russia and neighbouring countries: Identification of intertypic recombination in the S1 gene. Avian Pathol. 2011, 40, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Franzo, G.; Massi, P.; Tosi, G.; Blanco, A.; Antilles, N.; Biarnes, M.; Majo, N.; Nofrarias, M.; Dolz, R.; et al. A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 2017, 46, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Valastro, V.; Monne, I.; Fasolato, M.; Cecchettin, K.; Parker, D.; Terregino, C.; Cattoli, G. QX-type infectious bronchitis virus in commercial flocks in the UK. Vet. Rec. 2010, 167, 865–866. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jiang, Y.; Low, S.; Wang, Z.; Nam, S.J.; Liu, W.; Kwang, J. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001, 45, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Terregino, C.; Mazzacan, E.; Castaldello, I.; Capua, I.; Bonci, M. Detection of Chinese Q1 strain of infectious bronchitis virus in Europe. Vet. Rec. 2011, 169, 212–213. [Google Scholar] [CrossRef] [PubMed]
- Gelb, J., Jr.; Ladman, B.S.; Pope, C.R.; Ruano, J.M.; Brannick, E.M.; Bautista, D.A.; Coughlin, C.M.; Preskenis, L.A. Characterization of nephropathogenic infectious bronchitis virus DMV/1639/11 recovered from Delmarva broiler chickens in 2011. Avian Dis. 2013, 57, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.B.; Resurreccion, R.S. Genotyping of newly isolated infectious bronchitis virus isolates from northeastern Georgia. Avian Dis. 2010, 54, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.A.; Brash, M.L.; Hoyland, S.K.; Coventry, J.M.; Sandrock, C.; Guerin, M.T.; Ojkic, D. Genotyping of infectious bronchitis viruses identified in Canada between 2000 and 2013. Avian Pathol. 2014, 43, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.Y.; Sandri, T.L.; Souza, S.P.; Richtzenhain, L.J.; de Wit, J.J.; Brandao, P.E. Molecular epidemiology of avian infectious bronchitis in Brazil from 2007 to 2008 in breeders, broilers, and layers. Avian Dis. 2010, 54, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Chacon, J.L.; Rodrigues, J.N.; Assayag Junior, M.S.; Peloso, C.; Pedroso, A.C.; Ferreira, A.J. Epidemiological survey and molecular characterization of avian infectious bronchitis virus in Brazil between 2003 and 2009. Avian Pathol. 2011, 40, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Felippe, P.A.; da Silva, L.H.; Santos, M.M.; Spilki, F.R.; Arns, C.W. Genetic diversity of avian infectious bronchitis virus isolated from domestic chicken flocks and coronaviruses from feral pigeons in Brazil between 2003 and 2009. Avian Dis. 2010, 54, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Balestrin, E.; Fraga, A.P.; Ikuta, N.; Canal, C.W.; Fonseca, A.S.; Lunge, V.R. Infectious bronchitis virus in different avian physiological systems-a field study in Brazilian poultry flocks. Poult. Sci. 2014, 93, 1922–1929. [Google Scholar] [CrossRef] [PubMed]
- Marandino, A.; Pereda, A.; Tomas, G.; Hernandez, M.; Iraola, G.; Craig, M.I.; Hernandez, D.; Banda, A.; Villegas, P.; Panzera, Y.; et al. Phylodynamic analysis of avian infectious bronchitis virus in South America. J. Gen. Virol. 2015, 96, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Hewson, K.A.; Ignjatovic, J.; Browning, G.F.; Devlin, J.M.; Noormohammadi, A.H. Infectious bronchitis viruses with naturally occurring genomic rearrangement and gene deletion. Arch. Virol. 2011, 156, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Hilt, D.A.; Williams, S.M.; Woolcock, P.; Cardona, C.; O’Connor, R. Molecular and serologic characterization, pathogenicity, and protection studies with infectious bronchitis virus field isolates from California. Avian Dis. 2007, 51, 527–533. [Google Scholar] [CrossRef]
- Lee, C.-W.; Hilt, D.A.; Jackwood, M.W. Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene. J. Vet. Diagn. Investig. 2003, 15, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Dickhout, A.; Kutter, J.; Maier, H.J.; Britton, P.; Koumans, J.; Pijlman, G.P.; Fros, J.J.; Wiegertjes, G.F.; Forlenza, M. Infectious bronchitis coronavirus inhibits STAT1 signaling and requires accessory proteins for resistance to type I interferon activity. J. Virol. 2015, 89, 12047–12057. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Langereis, M.A.; Maier, H.J.; Britton, P.; van Kuppeveld, F.J.; Koumans, J.; Wiegertjes, G.F.; Forlenza, M. Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J. Virol. 2016, 90, 7519–7528. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Fernandez-Gutierrez, M.; Maier, H.J.; Britton, P.; Langereis, M.A.; Koumans, J.; Wiegertjes, G.F.; Forlenza, M. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J. Virol. 2015, 89, 1156–1167. [Google Scholar] [CrossRef] [PubMed]


| IBV Strain | Genotype | Lineage * | Geographic Origin | Immunization History | Arose From | Serotype or Protectotype | Viral Distribution | Mortality | References |
|---|---|---|---|---|---|---|---|---|---|
| CK/CH/LGX/111119 | Novel | GI-28 | China | H120 and 4/91 vaccinated yellow broiler | LX4, LDL/091022 and an unknown strain | Novel, different from Mass, 4/91, LX4, LDL/97I, and TW-I | Kidney, bronchi, cecal tonsils, oviduct | 30% in one-day-old specific-pathogen-free (SPF) white leghorn chickens with 106 50% embryo infective dose (EID50) | [20] |
| CK/CH/2010/JT-1 | Novel | GI-28 | China | H120 and 4/91 vaccinated broiler | QX, LSC/ 99I, LDT3/03 and 4/91 | Novel, different from Mass and 4/91 | Tracheal, lung, kidney | 43.75% in three-day-old SPF white leghorn chickens with 105 EID50 | [21] |
| SAIBK2 | Grouped in the most predominant genotype in China with YN, Sczy3, etc. | GI-19 | China | H120 vaccinated layer chickens | YX10, YN, and Mass | N.D.** | Kidney | 50% in one-day-old SPF chickens with 105 EID50 | [22] |
| CK/CH/LDL/110931 CK/CH/LHB/130573 | Mass | GI-1 | China | H120 vaccinated | H120-like vaccine strain and other types of viruses | Novel, different from H120, Conn and LDT3 | N.D. | N.D. | [23] |
| YN | Novel | GI-22 | China | H120 vaccinated broiler | N.D. | N.D. | Tracheal, lung, kidney, bursa, ovary, oviduct | 65% in 30-day-old SPF chickens with 105 EID50 40.5% in 21-day-old commercial laying hens with 106.5 EID50 | [24,25] |
| GD | TW-like | GI-7 | China | Mass-type vaccinated broiler | QX-like (S2, E, M and N) and TW-like (S1) | QX | Tracheal, lung, kidney, bursa, spleen, proventriculus | 40% in 21-day-old SPF chickens with 106 EID50 | [26,27] |
| CK/CH/LDL/140520 | TW-1 | GI-7 | China | H120 vaccinated | LX4 and Taiwan group 1 (TW-1) | Different from Mass, LDT3, 4/91, and Conn | Tracheal, kidney, oviduct | 30% in one-day-old SPF chickens with 105.5 EID50 | [28] |
| CK/CH/LGX/130530 | LDT3 | Unknown | China | H120 and Ma5 vaccinated broiler | H120 (nsp1-14) and LDT3/03 (nsp14-N) | LDT3-A vaccine provide protection while H120 not | Tracheal, kidney | 0% in one-day-old SPF chickens with 105 EID50 | [29] |
| CK/CH/LJL/111054 | Conn | GI-1 | China | H120 vaccinated layer | Mass type and Conn type | N.D. | N.D. | 0% in one-day-old SPF chickens with 105.5 EID50 | [30] |
| CK/CH/IBTZ/2012 | N.D. | GVI-1 | China | H120 vaccinated layer | LX4 and an unknown strain in S gene | N.D. | N.D. | N.D. | [31] |
| Sczy3 | LX4 | GI-19 | China | Unknown | LX4 (major) and H120 (minor) | N.D. | N.D. | N.D. | [32] |
| SAIBK | Novel | GI-22 | China | Unknown | SC021202 (major) and H120 | 4/91-like | [33] | ||
| 3575/08 | TW-I | GI-7 | Taiwan | TW-I vaccinated broiler | TW-I strain with point mutations | Novel, distinct with Mass or other local types | Trachea, proventriculus, kidney, bursa, oviduct | 83.3% in one-day-old SPF chickens with 106 EID50 | [34] |
| JP/Wakayama/2003 JP/Iwate/2005 | 4/91 | GI-13 | Japan | Without 4/91 immunization | N.D. | 4/91, JP/Iwate/2005 can be protected by 4/91 and JP-II strain. | Respiratory tract | 15.4% in one-day-old SPF chickens with 104 EID50 of JP/Wakayama/2003 | [35] |
| SNU8067 | Korean group I | GI-15 | South Korea | Unknown | Korean group II strain KM91 (except S1, 3a,3b) and Korean group I (S1) | KM91 and M41 inactivate vaccine protect 70% from oviduct lesion | Oviduct | N.D. | [36] |
| IBV/MN IBV/RA IBV/TU | Italy 02 | Unknown | Morocco | Unknown | N.D. | N.D. | Respiratory sign without renal lesion | 0% in one-day-old SPF chickens with 103.5 EID50 | [37] |
| IBV-EG/1212B-2012 IBV-EG/IBV1-2011 | Egy/Var II Egy/Var I | Unknown | Egypt | Unknown | Egyptian variant strains or Middle East strains with Israeli strains | N.D. | Tracheal, kidney | 50% to 40% in one-day-old SPF chickens with 105 EID50 | [38,39] |
| IBV/Cal99 variant/07 | Respiratory pathogenic Cal99 genotype | GI-9 | US | Game chickens without IBV vaccination | N.D. | N.D. | Trachea, lung, kidney, salivary glands, small intestine | N.D. | [40] |
| IBV/Brazil/2005/USP-10 | Brazilian type | Unknown | Brazil | Mass type vaccinated broiler | N.D. | N.D. | Respiratory tissue, kidney, intestine and testis | None in 26-day-old SPF chickens and male Ross broilers | [41] |
| γCoV/CK/Italy/I2022/13 IBV/FARM 1 IBV/FARM 5 | Q1 | GI-16 | Italy | H120 vaccinated broiler | Q1 and unknown minor parent strains | Q1 type | Trachea, lungs, kidneys, proventriculus | N.D., Ranging from 4.1% to 9.8% in broiler outbreak | [42,43] |
| KG3P | QX-like | GI-19 | England | 793B vaccinated Broiler | N.D. | N.D. | Trachea, lungs, kidneys, proventriculus | 16.7% in SPF chickens with 104 TOC50 | [44] |
| N1/08 | Australian group 2 | GIII-1 | Australia | Unknown | Australian group 2 and group 3 strains in S | N.D. | Trachea, cecal tonsils | None in 14-day-old SPF chickens with 105.5 EID50 | [45,46] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.-Y.; Chen, H.-W. Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation. Int. J. Mol. Sci. 2017, 18, 2030. https://doi.org/10.3390/ijms18102030
Lin S-Y, Chen H-W. Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation. International Journal of Molecular Sciences. 2017; 18(10):2030. https://doi.org/10.3390/ijms18102030
Chicago/Turabian StyleLin, Shu-Yi, and Hui-Wen Chen. 2017. "Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation" International Journal of Molecular Sciences 18, no. 10: 2030. https://doi.org/10.3390/ijms18102030