Bisphenol A Disrupts Transcription and Decreases Viability in Aging Vascular Endothelial Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bisphenol A (BPA) Alters Global Transcription in HT29 Human Colon Adenocarcinona Cell Line and Human Umbilical Vein Endothelial Cells (HUVEC)

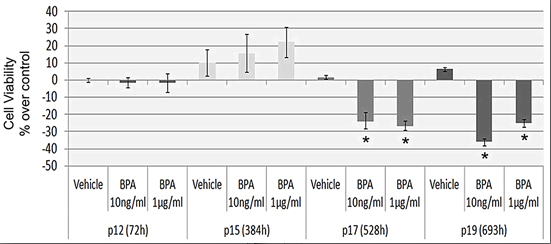
2.2. Continuous BPA Exposure Reduces Viability in Aging HUVEC

2.3. Continuous BPA Exposure Induces Differential Gene Expression in Aging HUVEC

3. Experimental Section
3.1. Cell Cultures and Reagents
3.2. BPA Treatments and Controls
3.3. cDNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
| Sequence | Accession No. | Forward Primer (5'→3') | Reverse Primer (5'→3') |
|---|---|---|---|
| L1-5' [24] | M80340.1 | GGCCAGTGTGTGTGCGCACCG | CCAGGTGTGGGATATAGTCTCGTGG |
| L1-3' [24] | M80340.1 | CAGGAAGGGGAATATCACACTC | TGCGCTGCACCCACTAACTC |
| SPARC | NM_003118 | CTGTGGGAGCTAATCCTG | GGGTGCTGGTCCAGCTGG |
| FN1 | NM_002026 | TGTGGTTGCCTTGCACGAT | GCTTGTGGGTGTGACCTGAGT |
| p21 | NM_000389 | CTGGAGACTCTCAGGGTCGAA | CCAGGACTGCAGGCTTCCT |
| FOS | NM_005252 | AGGAGAATCCGAAGGGAAAG | CAAGGGAAGCCACAGACATC |
| bcl-xL | NM_001191 | TTACCTGAATGACCACCTA | ATTTCCGACTGAAGAGTGA |
| NCL | NM_005381 | CCTTCTGAGGACATTCCAAGACA | ACGGTATTGCCCTTGAAATGTT |
| 18SrRNA | NR_003286 | CATTCGAACGTCTGCCCTAT | CCTCCAATGGATCCTCGTTA |
| GAPDH | NM_002046 | GAGTCAACGGATTTGGTCGTA | GCAGAGATGATGACCCTTTTG |
| β-actin | NM_001101 | GGTCATCTTCTCGCGGTTGGCCTTGGGGT | CCCCAGGCACCAGGGCGTGAT |
3.4. Protein Extraction and Western Blotting Analysis
3.5. Cell Viability Assay
3.6. Immunofluorescence
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mezcua, M.; Martínez-Uroz, M.A.; Gómez-Ramos, M.M.; Gómez, M.J.; Navas, J.M.; Fernández-Alba, A.R. Analysis of synthetic endocrine-disrupting chemicals in food: A review. Talanta 2012, 100, 90–106. [Google Scholar]
- Ballesteros-Gomez, A.; Rubio, S.; Pérez-Bendito, D. Analytical methods for the determination of bisphenol A in food. J. Chromatogr. A 2009, 1216, 449–469. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Chahoud, I.; Heindel, J.J.; Padmanabhan, V.; Paumgartten, F.J.; Schoenfelder, G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 2010, 118, 1055–1070. [Google Scholar] [CrossRef]
- Li, D.; Zhou, Z.; Qing, D.; He, Y.; Wu, T.; Miao, M.; Wang, J.; Weng, X.; Ferber, J.R.; Herrinton, L.J.; et al. Occupational exposure to bisphenol-A (BPA) and the risk of self-reported male sexual dysfunction. Hum. Reprod. 2010, 25, 519–527. [Google Scholar]
- He, Y.; Miao, M.; Wu, C.; Yuan, W.; Gao, E.; Zhou, Z.; Li, D.K. Occupational exposure levels of bisphenol A among Chinese workers. J. Occup. Health 2009, 51, 432–436. [Google Scholar]
- Rubin, B.S. Bisphenol A: An endocrine disruptor with widespread exposure and multiple effects. J. Steroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar]
- Ribeiro-Varandas, E.; Viegas, W.; Pereira, H.P.; Delgado, M. Bisphenol A at concentrations found in human serum induces aneugenic effects in endothelial cells. Mutat. Res. 2013, 751, 27–33. [Google Scholar] [CrossRef]
- Ptak, A.; Wróbel, A.; Gregoraszczuk, E.L. Effect of bisphenol-A on the expression of selected genes involved in cell cycle and apoptosis in the OVCAR-3 cell line. Toxicol. Lett. 2011, 202, 30–35. [Google Scholar]
- Singh, S.; Li, S.S. Epigenetic effects of environmental chemicals bisphenol a and phthalates. Int. J. Mol. Sci. 2012, 13, 10143–10153. [Google Scholar]
- Dolinoy, D.C.; Huang, D.; Jirtle, R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA 2007, 104, 13056–13061. [Google Scholar]
- Yaoi, T.; Itoh, K.; Nakamura, K.; Ogi, H.; Fujiwara, Y.; Fushiki, S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem. Biophys. Res. Commun. 2008, 376, 563–567. [Google Scholar]
- Fernandez, S.V.; Huang, Y.; Snider, K.E.; Zhou, Y.; Pogash, T.J.; Russo, J. Expression and DNA methylation changes in human breast epithelial cells after bisphenol A exposure. Int. J. Oncol. 2012, 41, 369–377. [Google Scholar]
- Doherty, L.F.; Bromer, J.G.; Zhou, Y.; Aldad, T.S.; Taylor, H.S. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: An epigenetic mechanism linking endocrine disruptors to breast cancer. Horm. Cancer 2010, 1, 146–155. [Google Scholar]
- Ribeiro-Varandas, E.; Pereira, H.S.; Monteiro, S.; Boavida Ferreira, R.; Neves, E.; Brito, L.; Viegas, W.; Delgado, M. The environmental pollutant bisphenol A interferes with nucleolar structure. In Proceedings of the 2012 International Conference on Biomedical Engineering and Biotechnology, Macau, China, 28–30 May 2012; IEEE Computer Society: Washington, DC, USA, 2012; pp. 1811–1814. [Google Scholar]
- D’Aquila, P.; Rose, G.; Bellizzi, D.; Passarino, G. Epigenetics and aging. Maturitas 2013, 74, 130–136. [Google Scholar]
- Boyd-Kirkup, J.D.; Green, C.D.; Wu, G.; Wang, D.; Han, J.D. Epigenomics and the regulation of aging. Epigenomics 2013, 5, 205–227. [Google Scholar]
- Dumont, P.; Chainiaux, F.; Eliaers, F.; Petropoulou, C.; Remacle, J.; Koch-Brandt, C.; Gonos, E.S.; Toussaint, O. Overexpression of apolipoprotein J in human fibroblasts protects against cytotoxicity and premature senescence induced by ethanol and tert-butylhydroperoxide. Cell Stress Chaperones 2002, 7, 23–35. [Google Scholar]
- Toussaint, O.; Medrano, E.E.; von Zglinicki, T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp. Gerontol. 2000, 35, 927–945. [Google Scholar]
- Qin, X.Y.; Fukuda, T.; Yang, L.; Zaha, H.; Akanuma, H.; Zeng, Q.; Yoshinaga, J.; Sone, H. Effects of bisphenol A exposure on the proliferation and senescence of normal human mammary epithelial cells. Cancer Biol. Ther. 2012, 13, 296–306. [Google Scholar]
- Melzer, D.; Gates, P.; Osborne, N.J.; Henley, W.E.; Cipelli, R.; Young, A.; Money, C.; McCormack, P.; Schofield, P.; Mosedale, D.; et al. Urinary bisphenol a concentration and angiography-defined coronary artery stenosis. PLoS One 2012, 7, e43378. [Google Scholar] [CrossRef] [Green Version]
- Melzer, D.; Rice, N.E.; Lewis, C.; Henley, W.E.; Galloway, T.S. Association of urinary bisphenol a concentration with heart disease: Evidence from NHANES 2003/06. PLoS One 2010, 5, e8673. [Google Scholar]
- Lind, P.M.; Lind, L. Circulating levels of bisphenol A and phthalates are related to carotid atherosclerosis in the elderly. Atherosclerosis 2011, 218, 207–213. [Google Scholar]
- Andreassi, M.G. DNA damage, vascular senescence and atherosclerosis. J. Mol. Med. (Berl.) 2008, 86, 1033–1043. [Google Scholar] [CrossRef]
- Aporntewan, C.; Phokaew, C.; Piriyapongsa, J.; Ngamphiw, C.; Ittiwut, C.; Tongsima, S.; Mutirangura, A. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One 2011, 6, e17934. [Google Scholar]
- Bredhult, C.; Sahlin, L.; Olovsson, M. Gene expression analysis of human endometrial endothelial cells exposed to Bisphenol A. Reprod. Toxicol. 2009, 28, 18–25. [Google Scholar]
- Naciff, J.M.; Khambatta, Z.S.; Reichling, T.D.; Carr, G.J.; Tiesman, J.P.; Singleton, D.W.; Khan, S.A.; Daston, G.P. The genomic response of Ishikawa cells to bisphenol A exposure is dose- and time-dependent. Toxicology 2010, 270, 137–149. [Google Scholar]
- Buterin, T.; Koch, C.; Naegeli, H. Convergent transcriptional profiles induced by endogenous estrogen and distinct xenoestrogens in breast cancer cells. Carcinogenesis 2006, 27, 1567–1578. [Google Scholar]
- Boehme, K. Simon, S.; Mueller, S.O. Gene expression profiling in Ishikawa cells: A fingerprint for estrogen active compounds. Toxicol. Appl. Pharmacol. 2009, 236, 85–96. [Google Scholar] [CrossRef]
- Ostertag, E.M.; Kazazian, H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001, 35, 501–538. [Google Scholar]
- Hon, G.; Wang, W.; Ren, B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput. Biol. 2009, 5, e1000566. [Google Scholar]
- Varier, R.A.; Timmers, H.T. Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 2011, 1815, 75–89. [Google Scholar]
- Dong, S.; Terasaka, S.; Kiyama, R. Bisphenol A induces a rapid activation of Erk1/2 through GPR30 in human breast cancer cells. Environ. Pollut. 2011, 159, 212–218. [Google Scholar]
- Pupo, M.; Pisano, A.; Lappano, R.; Santolla, M.F.; de Francesco, E.M.; Abonante, S.; Rosano, C.; Maggiolini, M. Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ. Health Perspect. 2012, 120, 1177–1182. [Google Scholar]
- Hanahan, D.; RWeinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Khaidakov, M.; Wang, X.; Mehta, J.L. Potential involvement of LOX-1 in functional consequences of endothelial senescence. PLoS One 2011, 6, e20964. [Google Scholar]
- Hwang, E.S.; Yoon, G.; Kang, H.T. A comparative analysis of the cell biology of senescence and aging. Cell Mol. Life Sci. 2009, 66, 2503–2524. [Google Scholar]
- Wagner, M.; Hampel, B.; Bernhard, D.; Hala, M.; Zwerschke, W.; Jansen-Dürr, P. Replicative senescence of human endothelial cells in vitro involves G1 arrest, polyploidization and senescence-associated apoptosis. Exp. Gerontol. 2001, 36, 1327–1347. [Google Scholar] [CrossRef]
- Wang, E.; Gundersen, D. Increased organization of cytoskeleton accompanying the aging of human fibroblasts in vitro. Exp. Cell Res. 1984, 154, 191–202. [Google Scholar]
- Van der Loo, B.; Fenton, M.J.; Erusalimsky, J.D. Cytochemical detection of a senescence-associated β-galactosidase in endothelial and smooth muscle cells from human and rabbit blood vessels. Exp. Cell Res. 1998, 241, 309–315. [Google Scholar]
- George, O.; Bryant, B.K.; Chinnasamy, R.; Corona, C.; Arterburn, J.B.; Shuster, C.B. Bisphenol A directly targets tubulin to disrupt spindle organization in embryonic and somatic cells. ACS Chem. Biol. 2008, 3, 167–179. [Google Scholar]
- Rivera, L.B.; Bradshaw, A.D.; Brekken, R.A. The regulatory function of SPARC in vascular biology. Cell Mol. Life Sci. 2011, 68, 3165–3173. [Google Scholar]
- Astrof, S.; Hynes, R.O. Fibronectins in vascular morphogenesis. Angiogenesis 2009, 12, 165–175. [Google Scholar]
- Debacq-Chainiaux, F.; Pascal, T.; Boilan, E.; Bastin, C.; Bauwens, E.; Toussaint, O. Screening of senescence-associated genes with specific DNA array reveals the role of IGFBP-3 in premature senescence of human diploid fibroblasts. Free Radic. Biol. Med. 2008, 44, 1817–1832. [Google Scholar]
- Puzianowska-Kuznicka, M.; Kuznicki, J. Genetic alterations in accelerated ageing syndromes. Do they play a role in natural ageing? Int. J. Biochem. Cell Biol. 2005, 37, 947–960. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar]
- De Camé Trécesson, S.; Guillemin, Y.; Bélanger, A.; Bernard, A.C.; Preisser, L.; Ravon, E.; Gamelin, E.; Juin, P.; Barré, B.; Coqueret, O. Escape from p21-mediated oncogene-induced senescence leads to cell dedifferentiation and dependence on anti-apoptotic Bcl-xL and MCL1 proteins. J. Biol. Chem. 2011, 286, 12825–12838. [Google Scholar]
- Kim, J.S.; Lee, S.; Lee, T.; Lee, Y.W.; Trepel, J.B. Transcriptional activation of p21(WAF1/CIP1) by apicidin, a novel histone deacetylase inhibitor. Biochem. Biophys. Res. Commun. 2001, 281, 866–871. [Google Scholar]
- Michels, J.; Kepp, O.; Senovilla, L.; Lissa, D.; Castedo, M.; Kroemer, G.; Galluzzi, L. Functions of bcl-xl at the interface between cell death and metabolism. Int. J. Cell Biol. 2013, 2013, 705294. [Google Scholar]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar]
- Higashi, Y.; Sukhanoy, S.; Anwar, A.; Shai, S.; Delafontaine, P. Aging, atherosclerosis, and IGF-1. J. Gerontol. A 2012, 67A, 626–639. [Google Scholar]
- Van Vre, E.A.; Ait-Oufella, H.; Tedgui, A.; Mallat, Z. Apoptotic cell death and efferocytosis in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 887–893. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ribeiro-Varandas, E.; Pereira, H.S.; Monteiro, S.; Neves, E.; Brito, L.; Ferreira, R.B.; Viegas, W.; Delgado, M. Bisphenol A Disrupts Transcription and Decreases Viability in Aging Vascular Endothelial Cells. Int. J. Mol. Sci. 2014, 15, 15791-15805. https://doi.org/10.3390/ijms150915791
Ribeiro-Varandas E, Pereira HS, Monteiro S, Neves E, Brito L, Ferreira RB, Viegas W, Delgado M. Bisphenol A Disrupts Transcription and Decreases Viability in Aging Vascular Endothelial Cells. International Journal of Molecular Sciences. 2014; 15(9):15791-15805. https://doi.org/10.3390/ijms150915791
Chicago/Turabian StyleRibeiro-Varandas, Edna, H. Sofia Pereira, Sara Monteiro, Elsa Neves, Luísa Brito, Ricardo Boavida Ferreira, Wanda Viegas, and Margarida Delgado. 2014. "Bisphenol A Disrupts Transcription and Decreases Viability in Aging Vascular Endothelial Cells" International Journal of Molecular Sciences 15, no. 9: 15791-15805. https://doi.org/10.3390/ijms150915791