Consequences of Hybridization in Mammals: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. Hybridization in Mammalian Orders and Families
4.1.1. Mammalian Orders
4.1.2. Mammalian Families
4.2. Typical Outcomes of Hybridization between Mammalian Species
4.2.1. Negative Outcomes
Genetic Swamping
Extinction via Genetic Swamping
Outbreeding Depression
Introgression from a Domesticated Lineage
Morphological Anomalies
Loss of Reproductive Output
4.2.2. Positive Outcomes
Increase in Genetic Diversity and Reduction of Inbreeding
Novel Adaptive Variation
Hybrid Speciation
4.2.3. Neutral or Unspecified Outcomes
Intermediate Phenotypic Traits
Hybrid Zones
Hybridization without Significant Impacts
No or Rare Evidence of Hybridization
4.2.4. Consequences of Hybridization for Threatened Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayr, E. Animal Species and Evolution; Belknap: Cambridge, MA, USA, 1963. [Google Scholar]
- Taylor, S.A.; Larson, E. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 2019, 3, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L. Natural Hybridization and Evolution; Oxford University Press on Demand: Oxford, UK, 1997. [Google Scholar]
- Nesi, N.; Nakoune, E.; Cruaud, C.; Hassanin, A. DNA barcoding of African fruit bats (Mammalia, Pteropodidae). The mitochondrial genome does not provide a reliable discrimination between Epomophorus Gamb and Micropteropus pusillus. C. R. Biol. 2011, 334, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.; Barton, N.H.; Good, J.M. Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 2016, 25, 2325–2332. [Google Scholar] [CrossRef] [Green Version]
- Goulet, B.E.; Roda, F.; Hopkins, R. Hybridization in Plants: Old Ideas, New Techniques. Plant Physiol. 2017, 173, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wayne, R.K.; Shaffer, H.B. Hybridization and endangered species protection in the molecular era. Mol. Ecol. 2016, 25, 2680–2689. [Google Scholar] [CrossRef]
- Mota, M.R.; Pinheiro, F.; Leal, B.S.S.; Wendt, T.; Palma-Silva, C. The role of hybridization and introgression in maintaining species integrity and cohesion in naturally isolated inselberg bromeliad populations. Plant Biol. 2019, 21, 122–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, A.C.; Woodward, G.; Seehausen, O.; Muñoz-Fuentes, V.; Moritz, C.; Guelmami, A.; Abbott, R.J.; Edelaar, P. Hybridization due to changing species distributions: Adding problems or solutions to conservation of biodiversity during global change? Evol. Ecol. Res. 2015, 16, 475–491. [Google Scholar]
- Lavrenchenko, L.A.; Bulatova, N.S. The role of hybrid zones in speciation: A case study on chromosome races of the house mouse Mus domesticus and common shrew Sorex araneus. Biol. Bull. Rev. 2016, 6, 232–244. [Google Scholar] [CrossRef]
- Chan, W.Y.; Peplow, L.M.; Menéndez, P.; Hoffmann, A.; van Oppen, M. Interspecific Hybridization May Provide Novel Opportunities for Coral Reef Restoration. Front. Mar. Sci. 2018, 5, 160. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, J.A.; Miller, J. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgro, C. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef]
- Olano-Marin, J.; Plis, K.; Sönnichsen, L.; Borowik, T.; Niedziałkowska, M.; Jędrzejewska, B. Weak population structure in European roe deer (Capreolus capreolus) and evidence of introgressive hybridization with Siberian roe deer (C. pygargus) in northeastern Poland. PLoS ONE 2014, 9, e109147. [Google Scholar]
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [Green Version]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef]
- Lavrenchenko, L. Hybrid speciation in mammals: Illusion or reality? Biol. Bull. Rev. 2014, 4, 198–209. [Google Scholar] [CrossRef]
- Canu, A.; Vilaça, S.; Iacolina, L.; Apollonio, M.; Bertorelle, G.; Scandura, M. Lack of polymorphism at the MC1R wild-type allele and evidence of domestic allele introgression across European wild boar populations. Mamm. Biol. 2016, 81, 477–479. [Google Scholar] [CrossRef]
- Zemanová, B.; Hájková, P.; Hájek, B.; Martínková, N.; Mikulíček, P.; Zima, J.; Bryja, J. Extremely low genetic variation in endangered Tatra chamois and evidence for hybridization with an introduced Alpine population. Conserv. Genet. 2015, 16, 729–741. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Pinedo-Castro, M.; Shostell, J.M. Small spotted bodies with multiple specific mitochondrial DNAs: Existence of diverse and differentiated tigrina lineages or species (Leopardus spp.: Felidae, Mammalia) throughout Latin America. Mitochondrial DNA Part A 2018, 29, 993–1014. [Google Scholar] [CrossRef]
- Draper, D.; Laguna, E.; Marques, I. Demystifying Negative Connotations of Hybridization for Less Biased Conservation Policies. Front. Ecol. Evol. 2021, 9, 268. [Google Scholar] [CrossRef]
- Cairns, K.M.; Nesbitt, B.J.; Laffan, S.W.; Letnic, M.; Crowther, M.S. Geographic hot spots of dingo genetic ancestry in southeastern Australia despite hybridisation with domestic dogs. Conserv. Genet. 2020, 21, 77–90. [Google Scholar] [CrossRef]
- Harris, K.; Nielsen, R. The Genetic Cost of Neanderthal Introgression. Genetics 2016, 203, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauclerc, K.B.; Bowman, J.; Schulte-Hostedde, A.I. Assessing the cryptic invasion of a domestic conspecific: A merican mink in their native range. Ecol. Evol. 2013, 3, 2296–2309. [Google Scholar] [CrossRef]
- Grobler, P.; van Wyk, A.M.; Dalton, D.L.; van Vuuren, B.J.; Kotzé, A. Assessing introgressive hybridization between blue wildebeest (Connochaetes taurinus) and black wildebeest (Connochaetes gnou) from South Africa. Conserv. Genet. 2018, 19, 981–993. [Google Scholar] [CrossRef]
- Koutsogiannouli, E.A.; Moutou, K.A.; Sarafidou, T.; Stamatis, C.; Mamuris, Z. Detection of hybrids between wild boars (Sus scrofa scrofa) and domestic pigs (Sus scrofa f. domestica) in Greece, using the PCR-RFLP method on melanocortin-1 receptor (MC1R) mutations. Mamm. Biol. 2010, 75, 69–73. [Google Scholar] [CrossRef]
- Colella, J.P.; Wilson, R.E.; Talbot, S.L.; Cook, J.A. Implications of introgression for wildlife translocations: The case of North American martens. Conserv. Genet. 2019, 20, 153–166. [Google Scholar] [CrossRef]
- Drygala, F.; Rode-Margono, J.; Semiadi, G.; Frantz, A.C. Evidence of hybridisation between the common Indonesian banded pig (Sus scrofa vitattus) and the endangered Java warty pig (Sus verrucosus). Conserv. Genet. 2020, 21, 1073–1078. [Google Scholar] [CrossRef]
- Haus, T.; Roos, C.; Zinner, D. Discordance between spatial distributions of Y-chromosomal and mitochondrial haplotypes in African green monkeys (Chlorocebus spp.): A result of introgressive hybridization or cryptic diversity? Int. J. Primatol. 2013, 34, 986–999. [Google Scholar] [CrossRef] [Green Version]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G. Hybridization and extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef]
- Balao, F.; Casimiro-Soriguer, R.; García-Castaño, J.L.; Terrab, A.; Talavera, S. Big thistle eats the little thistle: Does unidirectional introgressive hybridization endanger the conservation of Onopordum hinojense? New Phytol. 2015, 206, 448–458. [Google Scholar] [CrossRef]
- Lepais, O.; Petit, R.; Guichoux, E.; Lavabre, J.; Alberto, F.; Kremer, A.; Gerber, S. Species relative abundance and direction of introgression in oaks. Mol. Ecol. 2009, 18, 2228–2242. [Google Scholar] [CrossRef] [PubMed]
- Hirashiki, C.; Kareiva, P.; Marvier, M. Concern over hybridization risks should not preclude conservation interventions. Conserv. Sci. Pract. 2021, 3, e424. [Google Scholar] [CrossRef]
- Eva, S.N.; Yamazaki, Y. Hybridization between native and introduced individuals of sika deer in the central part of Toyama Prefecture. Mammal Study 2018, 43, 269–274. [Google Scholar] [CrossRef]
- Sagua, M.I.; Figueroa, C.; Acosta, D.; Fernández, G.; Carpinetti, B.; Birochio, D.; Merino, M.L. Inferring the origin and genetic diversity of the introduced wild boar (Sus scrofa) populations in Argentina: An approach from mitochondrial markers. Mammal Res. 2018, 63, 467–476. [Google Scholar] [CrossRef]
- Pilot, M.; Greco, C.; vonHoldt, B.M.; Randi, E.; Jędrzejewski, W.; Sidorovich, V.E.; Konopiński, M.K.; Ostrander, E.A.; Wayne, R.K. Widespread, long-term admixture between grey wolves and domestic dogs across Eurasia and its implications for the conservation status of hybrids. Evol. Appl. 2018, 11, 662–680. [Google Scholar] [CrossRef] [Green Version]
- Iacolina, L.; Scandura, M.; Gazzola, A.; Cappai, N.; Capitani, C.; Mattioli, L.; Vercillo, F.; Apollonio, M. Y-chromosome microsatellite variation in Italian wolves: A contribution to the study of wolf-dog hybridization patterns. Mamm. Biol. 2010, 75, 341–347. [Google Scholar] [CrossRef]
- Hulva, P.; Černá Bolfíková, B.; Woznicová, V.; Jindřichová, M.; Benešová, M.; Mysłajek, R.W.; Nowak, S.; Szewczyk, M.; Niedźwiecka, N.; Figura, M. Wolves at the crossroad: Fission–fusion range biogeography in the Western Carpathians and Central Europe. Divers. Distrib. 2018, 24, 179–192. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Fuentes, V.; Darimont, C.T.; Paquet, P.C.; Leonard, J.A. The genetic legacy of extirpation and re-colonization in Vancouver Island wolves. Conserv. Genet. 2010, 11, 547–556. [Google Scholar] [CrossRef] [Green Version]
- Santostasi, N.L.; Gimenez, O.; Caniglia, R.; Fabbri, E.; Molinari, L.; Reggioni, W.; Ciucci, P. Estimating Admixture at the Population Scale: Taking Imperfect Detectability and Uncertainty in Hybrid Classification Seriously. J. Wildl. Manag. 2021. [Google Scholar] [CrossRef]
- Cairns, K.M.; Newman, K.D.; Crowther, M.S.; Letnic, M. Pelage variation in dingoes across southeastern Australia: Implications for conservation and management. J. Zool. 2021, 314, 104–115. [Google Scholar] [CrossRef]
- Mattucci, F.; Oliveira, R.; Lyons, L.A.; Alves, P.C.; Randi, E. European wildcat populations are subdivided into five main biogeographic groups: Consequences of Pleistocene climate changes or recent anthropogenic fragmentation? Ecol. Evol. 2016, 6, 3–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beutel, T.; Reineking, B.; Tiesmeyer, A.; Nowak, C.; Heurich, M. Spatial patterns of co-occurrence of the European wildcat Felis silvestris silvestris and domestic cats Felis silvestris catus in the Bavarian Forest National Park. Wildl. Biol. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.; Randi, E.; Mattucci, F.; Kurushima, J.; Lyons, L.A.; Alves, P. Toward a genome-wide approach for detecting hybrids: Informative SNPs to detect introgression between domestic cats and European wildcats (Felis silvestris). Heredity 2015, 115, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Le Roux, J.J.; Foxcroft, L.C.; Herbst, M.; MacFadyen, S. Genetic analysis shows low levels of hybridization between A frican wildcats (Felis silvestris lybica) and domestic cats (F. s. catus) in South Africa. Ecol. Evol. 2015, 5, 288–299. [Google Scholar] [CrossRef]
- Boitani, L. Genetic considerations on wolf conservation in Italy. Ital. J. Zool. 1984, 51, 367–373. [Google Scholar] [CrossRef]
- Gottelli, D.; Sillero-Zubiri, C.; Applebaum, G.D.; Roy, M.S.; Girman, D.J.; Garcia-Moreno, J.; Ostrander, E.A.; Wayne, R.K. Molecular genetics of the most endangered canid: The Ethiopian wolf Canis simensis. Mol. Ecol. 1994, 3, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Randi, E. Genetics and conservation of wolves Canis lupus in Europe. Mammal Rev. 2011, 41, 99–111. [Google Scholar] [CrossRef]
- Torres, R.T.; Ferreira, E.; Rocha, R.G.; Fonseca, C. Hybridization between wolf and domestic dog: First evidence from an endangered population in central Portugal. Mamm. Biol. 2017, 86, 70–74. [Google Scholar] [CrossRef]
- Popova, E.; Zlatanova, D. Living a dog’s life: A putative gray wolf in a feral dog group. Mammalia 2020, 84, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Saetre, P.; Lindberg, J.; Leonard, J.A.; Olsson, K.; Pettersson, U.; Ellegren, H.; Bergström, T.F.; Vila, C.; Jazin, E. From wild wolf to domestic dog: Gene expression changes in the brain. Mol. Brain Res. 2004, 126, 198–206. [Google Scholar] [CrossRef]
- Lounsberry, Z.T.; Quinn, C.B.; Statham, M.J.; Angulo, C.L.; Kalani, T.J.; Tiller, E.; Sacks, B.N. Investigating genetic introgression from farmed red foxes into the wild population in Newfoundland, Canada. Conserv. Genet. 2017, 18, 383–392. [Google Scholar] [CrossRef]
- Arnold, M.L.; Martin, N.H. Hybrid fitness across time and habitats. Trends Ecol. Evol. 2010, 25, 530–536. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, S.E.; Pemberton, J.M. Detecting the true extent of introgression during anthropogenic hybridization. Trends Ecol. Evol. 2019, 34, 315–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shurtliff, Q.R. Mammalian hybrid zones: A review. Mammal Rev. 2013, 43, 1–21. [Google Scholar] [CrossRef]
- Petit, R.J.; Excoffier, L. Gene flow and species delimitation. Trends Ecol. Evol. 2009, 24, 386–393. [Google Scholar] [CrossRef]
- Iacolina, L.; Corlatti, L.; Buzan, E.; Safner, T.; Šprem, N. Hybridisation in European ungulates: An overview of the current status, causes, and consequences. Mammal Rev. 2019, 49, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Morgan-Richards, M.; Smissen, R.D.; Shepherd, L.D.; Wallis, G.P.; Hayward, J.J.; Chan, C.H.; Chambers, G.K.; Chapman, H.M. A review of genetic analyses of hybridisation in New Zealand. J. R. Soc. N. Z. 2009, 39, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, A.R.E. Mammal population regulation, keystone processes and ecosystem dynamics. Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 1729–1740. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.E.; Reeder, D.M. Mammal Species of the World: A Taxonomic and Geographic Reference; JHU Press: Baltimore, MD, USA, 2005; Volume 1. [Google Scholar]
- Tolesa, Z.; Bekele, E.; Tesfaye, K.; Ben Slimen, H.; Valqui, J.; Getahun, A.; Hartl, G.B.; Suchentrunk, F. Mitochondrial and nuclear DNA reveals reticulate evolution in hares (Lepus spp., Lagomorpha, Mammalia) from Ethiopia. PLoS ONE 2017, 12, e0180137. [Google Scholar] [CrossRef] [Green Version]
- Grobler, J.P.; Hayter, K.N.; Labuschagne, C.; Nel, E.; Coetzer, W.G. The genetic status of naturally occurring black-nosed impala from northern South Africa. Mamm. Biol. 2017, 82, 27–33. [Google Scholar] [CrossRef]
- Moura, A.E.; Tsingarska, E.; Dąbrowski, M.J.; Czarnomska, S.D.; Jędrzejewska, B.; Pilot, M. Unregulated hunting and genetic recovery from a severe population decline: The cautionary case of Bulgarian wolves. Conserv. Genet. 2014, 15, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Cairns, K.M.; Brown, S.K.; Sacks, B.N.; Ballard, J.W.O. Conservation implications for dingoes from the maternal and paternal genome: Multiple populations, dog introgression, and demography. Ecol. Evol. 2017, 7, 9787–9807. [Google Scholar] [CrossRef] [Green Version]
- Canu, A.; Apollonio, M.; Scandura, M. Unmasking the invader: Genetic identity of invasive wild boar from three minor islands off Sardinia (Italy). Mamm. Biol. 2018, 93, 29–37. [Google Scholar] [CrossRef]
- Artyushin, I.; Bannikova, A.; Lebedev, V.; Kruskop, S. Mitochondrial DNA relationships among North Palaearctic Eptesicus (Vespertilionidae, Chiroptera) and past hybridization between common serotine and northern bat. Zootaxa 2009, 2262, 40–52. [Google Scholar]
- Berthier, P.; Excoffier, L.; Ruedi, M. Recurrent replacement of mtDNA and cryptic hybridization between two sibling bat species Myotis myotis and Myotis blythii. Proc. R. Soc. B Biol. Sci. 2006, 273, 3101–3123. [Google Scholar] [CrossRef] [Green Version]
- Bogdanowicz, W.; Van Den Bussche, R.A.; Gajewska, M.; Postawa, T.; Harutyunyan, M. Ancient and contemporary DNA sheds light on the history of mouse-eared bats in Europe and the Caucasus. Acta Chiropterol. 2009, 11, 289–305. [Google Scholar] [CrossRef]
- Hoffmann, F.G.; Owen, J.G.; Baker, R.J. mtDNA perspective of chromosomal diversification and hybridization in Peters’ tent-making bat (Uroderma bilobatum: Phyllostomidae). Mol. Ecol. 2003, 12, 2981–2993. [Google Scholar] [CrossRef]
- Afonso, E.; Goydadin, A.-C.; Giraudoux, P.; Farny, G. Investigating hybridization between the two sibling bat species Myotis myotis and M. blythii from guano in a natural mixed maternity colony. PLoS ONE 2017, 12, e0170534. [Google Scholar] [CrossRef]
- Vallo, P.; Benda, P.; Červený, J.; Koubek, P. Conflicting mitochondrial and nuclear paraphyly in small-sized West African house bats (Vespertilionidae). Zool. Scr. 2013, 42, 1–12. [Google Scholar] [CrossRef]
- Bogdanowicz, W.; Piksa, K.; Tereba, A. Hybridization hotspots at bat swarming sites. PLoS ONE 2012, 7, e53334. [Google Scholar] [CrossRef] [Green Version]
- Arlettaz, R.; Christe, P.; Lugon, A.; Perrin, N.; Vogel, P. Food availability dictates the timing of parturition in insectivorous mouse-eared bats. Oikos 2001, 95, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Linnell, J.D.; Swenson, J.E.; Andersen, R. Conservation of biodiversity in Scandinavian boreal forests: Large carnivores as flagships, umbrellas, indicators, or keystones? Biodivers. Conserv. 2000, 9, 857–868. [Google Scholar] [CrossRef]
- Macdonald, E.; Burnham, D.; Hinks, A.; Dickman, A.; Malhi, Y.; Macdonald, D. Conservation inequality and the charismatic cat: Felis felicis. Glob. Ecol. Conserv. 2015, 3, 851–866. [Google Scholar] [CrossRef] [Green Version]
- Tensen, L. Biases in wildlife and conservation research, using felids and canids as a case study. Glob. Ecol. Conserv. 2018, 15, e00423. [Google Scholar] [CrossRef]
- Tisdell, C.; Nantha, H.S.; Wilson, C. Endangerment and likeability of wildlife species: How important are they for payments proposed for conservation? Ecol. Econ. 2007, 60, 627–633. [Google Scholar] [CrossRef]
- Nyhus, P.J. Human–wildlife conflict and coexistence. Annu. Rev. Environ. Resour. 2016, 41, 143–171. [Google Scholar] [CrossRef] [Green Version]
- Caniglia, R.; Fabbri, E.; Galaverni, M.; Milanesi, P.; Randi, E. Noninvasive sampling and genetic variability, pack structure, and dynamics in an expanding wolf population. J. Mammal. 2014, 95, 41–59. [Google Scholar] [CrossRef] [Green Version]
- Smeds, L.; Aspi, J.; Berglund, J.; Kojola, I.; Tirronen, K.; Ellegren, H. Whole-genome analyses provide no evidence for dog introgression in Fennoscandian wolf populations. Evol. Appl. 2021, 14, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Boggiano, F.; Ciofi, C.; Boitani, L.; Formia, A.; Grottoli, L.; Natali, C.; Ciucci, P. Detection of an East European wolf haplotype puzzles mitochondrial DNA monomorphism of the Italian wolf population. Mamm. Biol. 2013, 78, 374–378. [Google Scholar] [CrossRef]
- Pilot, M.; Moura, A.E.; Okhlopkov, I.M.; Mamaev, N.V.; Alagaili, A.N.; Mohammed, O.B.; Yavruyan, E.G.; Manaseryan, N.H.; Hayrapetyan, V.; Kopaliani, N. Global phylogeographic and admixture patterns in grey wolves and genetic legacy of an ancient Siberian lineage. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zwijacz-Kozica, T.; Ważna, A.; Muñoz-Fuentes, V.; Tiesmeyer, A.; Cichocki, J.; Nowak, C. Not European wildcats, but domestic cats inhabit Tatra National Park. Pol. J. Ecol. 2017, 65, 415–421. [Google Scholar] [CrossRef]
- Nussberger, B.; Wandeler, P.; Weber, D.; Keller, L.F. Monitoring introgression in European wildcats in the Swiss Jura. Conserv. Genet. 2014, 15, 1219–1230. [Google Scholar] [CrossRef] [Green Version]
- Leonard, J.A.; Echegaray, J.; Rand, E.; Vilà, C. Impact of hybridization with domestic dogs on the conservation of wild canids. Free Ranging Dogs Wildl. Conserv. 2013, 170, 170–184. [Google Scholar] [CrossRef]
- Ottoni, C.; Van Neer, W.; De Cupere, B.; Daligault, J.; Guimaraes, S.; Peters, J.; Spassov, N.; Prendergast, M.E.; Boivin, N.; Morales-Muñiz, A. The palaeogenetics of cat dispersal in the ancient world. Nat. Ecol. Evol. 2017, 1, 1–7. [Google Scholar] [CrossRef]
- Vigne, J.-D.; Guilaine, J.; Debue, K.; Haye, L.; Gérard, P. Early taming of the cat in Cyprus. Science 2004, 304, 259. [Google Scholar] [CrossRef] [Green Version]
- Gompper, M.E. The dog-human-wildlife interface: Assessing the scope of the problem. Free Ranging Dogs Wildl. Conserv. 2014, 9–54. [Google Scholar] [CrossRef]
- Adams, J.R.; Leonard, J.A.; Waits, L.P. Widespread occurrence of a domestic dog mitochondrial DNA haplotype in southeastern US coyotes. Mol. Ecol. 2003, 12, 541–546. [Google Scholar] [CrossRef]
- Vilà, C.; Maldonado, J.E.; Wayne, R.K. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. J. Hered. 1999, 90, 71–77. [Google Scholar] [CrossRef]
- Dzialuk, A.; Zastempowska, E.; Skórzewski, R.; Twarużek, M.; Grajewski, J. High domestic pig contribution to the local gene pool of free-living European wild boar: A case study in Poland. Mammal Res. 2018, 63, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Apollonio, M.; Andersen, R.; Putman, R. European Ungulates and Their Management in the Twenty-First Century; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Herrero, J.; García-Serrano, A.; García-González, R. Reproductive and demographic parameters in two Iberian wild boar Sus scrofa populations. Mammal Res. 2008, 53, 355–364. [Google Scholar] [CrossRef]
- Massei, G.; Kindberg, J.; Licoppe, A.; Gačić, D.; Šprem, N.; Kamler, J.; Baubet, E.; Hohmann, U.; Monaco, A.; Ozoliņš, J. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag. Sci. 2015, 71, 492–500. [Google Scholar] [CrossRef]
- Vetter, S.G.; Ruf, T.; Bieber, C.; Arnold, W. What is a mild winter? Regional differences in within-species responses to climate change. PLoS ONE 2015, 10, e0132178. [Google Scholar] [CrossRef]
- Waithman, J.D.; Sweitzer, R.A.; Van Vuren, D.; Drew, J.D.; Brinkhaus, A.J.; Gardner, I.A.; Boyce, W.M. Range expansion, population sizes, and management of wild pigs in California. J. Wildl. Manag. 1999, 63, 298–308. [Google Scholar] [CrossRef]
- Frantz, A.C.; Zachos, F.E.; Kirschning, J.; Cellina, S.; Bertouille, S.; Mamuris, Z.; Koutsogiannouli, E.A.; Burke, T. Genetic evidence for introgression between domestic pigs and wild boars (Sus scrofa) in Belgium and Luxembourg: A comparative approach with multiple marker systems. Biol. J. Linn. Soc. 2013, 110, 104–115. [Google Scholar] [CrossRef] [Green Version]
- García, G.; Vergara, J.; Lombardi, R. Genetic characterization and phylogeography of the wild boar Sus scrofa introduced into Uruguay. Genet. Mol. Biol. 2011, 34, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Chai, Z.X.; Xin, J.W.; Zhang, C.F.; Zhang, Q.; Li, C.; Zhu, Y.; Cao, H.W.; Wang, H.; Han, J.L.; Ji, Q.M. Whole-genome resequencing provides insights into the evolution and divergence of the native domestic yaks of the Qinghai–Tibet Plateau. BMC Evol. Biol. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Anderson, E.; Stebbins, G.L., Jr. Hybridization as an evolutionary stimulus. Evolution 1954, 8, 378–388. [Google Scholar] [CrossRef]
- Flores-Manzanero, A.; Valenzuela-Galván, D.; Cuarón, A.D.; Vázquez-Domínguez, E. Conservation genetics of two critically endangered island dwarf carnivores. Conserv. Genet. 2021, 1–15. [Google Scholar] [CrossRef]
- Baskin, Y. A Plague of Rats and Rubbervines: The Growing Threat of Species Invasions; Island Press: Washington, DC, USA, 2013. [Google Scholar]
- McNeely, J.A. The Great Reshuffling: Human Dimensions of Invasive Alien Species; IUCN: Gland, Switzerland, 2001. [Google Scholar]
- Wittenberg, R.; Cock, M.J. Invasive Alien Species: A Toolkit of Best Prevention and Management Practices; CABI: Egham, UK, 2001. [Google Scholar]
- Queirós, J.; Gortázar, C.; Alves, P.C. Deciphering anthropogenic effects on the genetic background of the Red deer in the Iberian Peninsula. Front. Ecol. Evol. 2020, 8, 147. [Google Scholar] [CrossRef]
- Csányi, S.; Carranza, J.; Pokorny, B.; Putman, R.; Ryan, M. Valuing ungulates in Europe. In Behaviour and Management of European Ungulates; Whittles: Dunbeath, UK, 2014; pp. 13–45. [Google Scholar]
- Lever, C. Naturalized Mammals of the World; Longman: Harlow, UK, 1985. [Google Scholar]
- Kalb, D.M.; Bowman, J.L. A complete history of the establishment of Japanese sika deer on the Delmarva Peninsula: 100 years post-introduction. Biol. Invasions 2017, 19, 1705–1713. [Google Scholar] [CrossRef]
- Krojerová-Prokešová, J.; Barančeková, M.; Kawata, Y.; Oshida, T.; Igota, H.; Koubek, P. Genetic differentiation between introduced Central European sika and source populations in Japan: Effects of isolation and demographic events. Biol. Invasions 2017, 19, 2125–2141. [Google Scholar] [CrossRef]
- Takagi, T.; Matsumoto, Y.; Koda, R.; Tamate, H.B. Bi-directional movement of deer between Tomogashima islands and the western part of the Kii Peninsula, Japan, with special reference to hybridization between the Japanese sika deer (Cervus nippon centralis) and the introduced exotic deer. Mammal Study 2020, 45, 133–141. [Google Scholar] [CrossRef]
- Świsłocka, M.; Czajkowska, M.; Matosiuk, M.; Saveljev, A.P.; Ratkiewicz, M.; Borkowska, A. No evidence for recent introgressive hybridization between the European and Siberian roe deer in Poland. Mamm. Biol. 2019, 97, 59–63. [Google Scholar] [CrossRef]
- Mucci, N.; Mattucci, F.; Randi, E. Conservation of threatened local gene pools: Landscape genetics of the Italian roe deer (Capreolus c. italicus) populations. Evol. Ecol. Res. 2012, 14, 897–920. [Google Scholar]
- Do Nascimento Schaurich, M.; Lopes, F.R.V.; de Oliveira, L.R. Hybridization phenomenon in cetacean and pinniped species. Neotrop. Biol. Conserv. 2012, 7, 199–209. [Google Scholar]
- Glover, K.A.; Kanda, N.; Haug, T.; Pastene, L.A.; Øien, N.; Seliussen, B.B.; Sørvik, A.G.; Skaug, H.J. Hybrids between common and Antarctic minke whales are fertile and can back-cross. BMC Genet. 2013, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Crossman, C.A.; Taylor, E.B.; Barrett-Lennard, L.G. Hybridization in the Cetacea: Widespread occurrence and associated morphological, behavioral, and ecological factors. Ecol. Evol. 2016, 6, 1293–1303. [Google Scholar] [CrossRef] [Green Version]
- Miralles, L.; Lens, S.; Rodriguez-Folgar, A.; Carrillo, M.; Martin, V.; Mikkelsen, B.; Garcia-Vazquez, E. Interspecific introgression in cetaceans: DNA markers reveal post-F1 status of a pilot whale. PLoS ONE 2013, 8, e69511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Sun, D.; Cao, Y.; Xiao, L.; Huang, X.; Ren, W.; Xu, S.; Yang, G. Extensive Interspecific Gene Flow Shaped Complex Evolutionary History and Underestimated Species Diversity in Rapidly Radiated Dolphins. J. Mamm. Evol. 2021, 1–15. [Google Scholar] [CrossRef]
- Bérubé, M.; Palsbøll, P.J. Hybridism. In Encyclopedia of Marine Mammals; Elsevier: Amsterdam, The Netherlands, 2018; pp. 496–501. [Google Scholar]
- Willis, P.M.; Crespi, B.J.; Dill, L.M.; Baird, R.W.; Hanson, M.B. Natural hybridization between Dall’s porpoises (Phocoenoides dalli) and harbour porpoises (Phocoena phocoena). Can. J. Zool. 2004, 82, 828–834. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.M.; Kopps, A.M.; Allen, S.J.; Bejder, L.; Littleford-Colquhoun, B.; Parra, G.J.; Cagnazzi, D.; Thiele, D.; Palmer, C.; Frere, C.H. Population differentiation and hybridisation of Australian snubfin (Orcaella heinsohni) and Indo-Pacific humpback (Sousa chinensis) dolphins in north-western Australia. PLoS ONE 2014, 9, e101427. [Google Scholar] [CrossRef] [Green Version]
- Pampoulie, C.; Gíslason, D.; Ólafsdóttir, G.; Chosson, V.; Halldórsson, S.D.; Mariani, S.; Elvarsson, B.Þ.; Rasmussen, M.H.; Iversen, M.R.; Daníelsdóttir, A.K. Evidence of unidirectional hybridization and second-generation adult hybrid between the two largest animals on Earth, the fin and blue whales. Evol. Appl. 2021, 14, 314–321. [Google Scholar] [CrossRef]
- Quilodrán, C.S.; Montoya-Burgos, J.I.; Currat, M. Harmonizing hybridization dissonance in conservation. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- Brekke, T.D.; Henry, L.A.; Good, J.M. Genomic imprinting, disrupted placental expression, and speciation. Evolution 2016, 70, 2690–2703. [Google Scholar] [CrossRef] [Green Version]
- van Wyk, A.M.; Kotzé, A.; Randi, E.; Dalton, D.L. A hybrid dilemma: A molecular investigation of South African bontebok (Damaliscus pygargus pygargus) and blesbok (Damaliscus pygargus phillipsi). Conserv. Genet. 2013, 14, 589–599. [Google Scholar] [CrossRef]
- Bozarth, C.A.; Hailer, F.; Rockwood, L.L.; Edwards, C.W.; Maldonado, J.E. Coyote colonization of northern Virginia and admixture with Great Lakes wolves. J. Mammal. 2011, 92, 1070–1080. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Elfring, A.; Barrett, R.; Combs, M.; Davies, T.; Munshi-South, J.; Millien, V. Admixture on the northern front: Population genomics of range expansion in the white-footed mouse (Peromyscus leucopus) and secondary contact with the deer mouse (Peromyscus maniculatus). Heredity 2017, 119, 447–458. [Google Scholar] [CrossRef]
- Brekke, T.D.; Good, J.M. Parent-of-origin growth effects and the evolution of hybrid inviability in dwarf hamsters. Evolution 2014, 68, 3134–3148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neaves, L.E.; Zenger, K.; Cooper, D.W.; Eldridge, M. Molecular detection of hybridization between sympatric kangaroo species in south-eastern Australia. Heredity 2010, 104, 502–512. [Google Scholar] [CrossRef]
- Templeton, A.R.; Hemmer, H.; Mace, G.; Seal, U.S.; Shields, W.M.; Woodruff, D.S. Local adaptation, coadaptation, and population boundaries. Zoo Biol. 1986, 5, 115–125. [Google Scholar] [CrossRef]
- Bell, D.A.; Robinson, Z.L.; Funk, W.C.; Fitzpatrick, S.W.; Allendorf, F.W.; Tallmon, D.A.; Whiteley, A.R. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 2019, 34, 1070–1079. [Google Scholar] [CrossRef]
- Mallil, K.; Justy, F.; Rueness, E.K.; Dufour, S.; Totis, T.; Bloch, C.; Baarman, J.; Amroun, M.; Gaubert, P. Population genetics of the African wolf (Canis lupaster) across its range: First evidence of hybridization with domestic dogs in Africa. Mamm. Biol. 2020, 100, 645–658. [Google Scholar] [CrossRef]
- Smith, S.L.; Senn, H.V.; Pérez-Espona, S.; Wyman, M.T.; Heap, E.; Pemberton, J.M. Introgression of exotic Cervus (nippon and canadensis) into red deer (Cervus elaphus) populations in Scotland and the English Lake District. Ecol. Evol. 2018, 8, 2122–2134. [Google Scholar] [CrossRef] [Green Version]
- Bifolchi, A.; Picard, D.; Lemaire, C.; Cormier, J.; Pagano, A. Evidence of admixture between differentiated genetic pools at a regional scale in an invasive carnivore. Conserv. Genet. 2010, 11, 1–9. [Google Scholar] [CrossRef]
- Zhigileva, O.N.; Uslamina, I.M.; Gimranov, D.O.; Chernova, A.A. Mitochondrial DNA markers for the study of introgression between the sable and the pine marten. Conserv. Genet. Resour. 2020, 12, 329–336. [Google Scholar] [CrossRef]
- Pilot, M.; Moura, A.E.; Okhlopkov, I.M.; Mamaev, N.V.; Manaseryan, N.H.; Hayrapetyan, V.; Kopaliani, N.; Tsingarska, E.; Alagaili, A.N.; Mohammed, O.B. Human-modified canids in human-modified landscapes: The evolutionary consequences of hybridisation for grey wolves and free-ranging domestic dogs. Evol. Appl. 2021, 14, 2433–2456. [Google Scholar] [CrossRef]
- Gómez, J.M.; González-Megías, A.; Lorite, J.; Abdelaziz, M.; Perfectti, F. The silent extinction: Climate change and the potential hybridization-mediated extinction of endemic high-mountain plants. Biodivers. Conserv. 2015, 24, 1843–1857. [Google Scholar] [CrossRef]
- La Morgia, V.; Venturino, E. Understanding hybridization and competition processes between hare species: Implications for conservation and management on the basis of a mathematical model. Ecol. Model. 2017, 364, 13–24. [Google Scholar] [CrossRef]
- Godwin, J.L.; Lumley, A.J.; Michalczyk, Ł.; Martin, O.Y.; Gage, M.J. Mating patterns influence vulnerability to the extinction vortex. Glob. Chang. Biol. 2020, 26, 4226–4239. [Google Scholar] [CrossRef] [PubMed]
- Soulé, M.; Gilpin, M.; Conway, W.; Foose, T. The millenium ark: How long a voyage, how many staterooms, how many passengers? Zoo Biol. 1986, 5, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Ralls, K.; Ballou, J.D.; Frankham, R. Inbreeding and outbreeding. In Encyclopedia of Biodiversity; Elsevier: Amsterdam, The Netherlands, 2001. [Google Scholar]
- Biedrzycka, A.; Solarz, W.; Okarma, H. Hybridization between native and introduced species of deer in Eastern Europe. J. Mammal. 2012, 93, 1331–1341. [Google Scholar] [CrossRef] [Green Version]
- Muhlfeld, C.C.; Kalinowski, S.T.; McMahon, T.E.; Taper, M.L.; Painter, S.; Leary, R.F.; Allendorf, F.W. Hybridization rapidly reduces fitness of a native trout in the wild. Biol. Lett. 2009, 5, 328–331. [Google Scholar] [CrossRef] [Green Version]
- Templeton, A.R. Coadaptation and outbreeding depression. In Conservation Biology: The Science of Scarcity and Diversity; Sinauer Associates: Sunderland, MA, USA, 1986; pp. 105–116. [Google Scholar]
- Piorno, V.; Villafuerte, R.; Branco, M.; Carneiro, M.; Ferrand, N.; Alves, P. Low persistence in nature of captive reared rabbits after restocking operations. Eur. J. Wildl. Res. 2015, 61, 591–599. [Google Scholar] [CrossRef]
- Berejikian, B.A.; Ford, M.J. Review of Relative Fitness of Hatchery and Natural Salmon. 2004. Available online: https://www.webapps.nwfsc.noaa.gov/assets/25/6429_02012005_154209_fitnesstm61final.pdf (accessed on 11 November 2021).
- Fraser, D. Understanding animal welfare. Acta Vet. Scand. 2008, 50, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiik, K.; Maran, T.; Nagl, A.; Ashford, K.; Tammaru, T. The causes of the low breeding success of European mink (Mustela lutreola) in captivity. Zoo Biol. 2013, 32, 387–393. [Google Scholar] [CrossRef]
- Maran, T.; Põdra, M.; Põlma, M.; Macdonald, D.W. The survival of captive-born animals in restoration programmes–Case study of the endangered European mink Mustela lutreola. Biol. Conserv. 2009, 142, 1685–1692. [Google Scholar] [CrossRef]
- Randi, E. Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 2008, 17, 285–293. [Google Scholar] [CrossRef]
- Wierzbicki, H.; Zatoń-Dobrowolska, M.; Mucha, A.; Moska, M. Insight into the Genetic Population Structure of Wild Red Foxes in Poland Reveals Low Risk of Genetic Introgression from Escaped Farm Red Foxes. Genes 2021, 12, 637. [Google Scholar] [CrossRef] [PubMed]
- Feulner, P.G.; Gratten, J.; Kijas, J.W.; Visscher, P.M.; Pemberton, J.M.; Slate, J. Introgression and the fate of domesticated genes in a wild mammal population. Mol. Ecol. 2013, 22, 4210–4221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossen, C.; Keller, L.; Biebach, I.; Consortium, I.G.G.; Croll, D. Introgression from domestic goat generated variation at the major histocompatibility complex of alpine ibex. PLoS Genet. 2014, 10, e1004438. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, R.R.; Brink, J.S.; Vrahimis, S.; De Klerk, B. Hybrid wildebeest (Artiodactyla: Bovidae) provide further evidence for shared signatures of admixture in mammalian crania. S. Afr. J. Sci. 2010, 106, 1–5. [Google Scholar] [CrossRef]
- Tobler, M.; Carson, E.W. Environmental variation, hybridization, and phenotypic diversification in Cuatro Ciénegas pupfishes. J. Evol. Biol. 2010, 23, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A. Developmental anomalies in Drosophila hybrids are apparently caused by loss of microchromosome. Heredity 1990, 64, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, R.R. Phenotypic traits of primate hybrids: Recognizing admixture in the fossil record. Evol. Anthropol. Issues News Rev. 2010, 19, 258–270. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics; Pearson Education India: London, UK, 1996. [Google Scholar]
- Stelkens, R.B.; Schmid, C.; Selz, O.; Seehausen, O. Phenotypic novelty in experimental hybrids is predicted by the genetic distance between species of cichlid fish. BMC Evol. Biol. 2009, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Tung, J.; Charpentier, M.J.; Mukherjee, S.; Altmann, J.; Alberts, S.C. Genetic effects on mating success and partner choice in a social mammal. Am. Nat. 2012, 180, 113–129. [Google Scholar] [CrossRef] [Green Version]
- Zorenko, T.A.; Atanasov, N.; Golenishchev, F.N. Behavioral differentiation and hybridization of the European and Asian forms of Harting’vole Microtus hartingi (Rodentia, Arvicolinae). Russ. J. Theriol. 2016, 15, 133–150. [Google Scholar] [CrossRef]
- Chan, W.Y.; Hoffmann, A.A.; van Oppen, M.J. Hybridization as a conservation management tool. Conserv. Lett. 2019, 12, e12652. [Google Scholar] [CrossRef]
- Whiteley, A.R.; Fitzpatrick, S.W.; Funk, W.C.; Tallmon, D.A. Genetic rescue to the rescue. Trends Ecol. Evol. 2015, 30, 42–49. [Google Scholar] [CrossRef]
- Ralls, K.; Sunnucks, P.; Lacy, R.C.; Frankham, R. Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 2020, 251, 108784. [Google Scholar] [CrossRef]
- Salzburger, W.; Baric, S.; Sturmbauer, C. Speciation via introgressive hybridization in East African cichlids? Mol. Ecol. 2002, 11, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, Z.; Aliabadian, M.; Ghorbani, F.; Moghaddam, F.Y.; Lissovsky, A.A.; Obst, M.; Olsson, U. Unidirectional Introgression and Evidence of Hybrid Superiority over Parental Populations in Eastern Iranian Plateau Population of Hares (Mammalia: Lepus Linnaeus, 1758). J. Mamm. Evol. 2020, 27, 723–743. [Google Scholar] [CrossRef]
- Chen, Z.-H.; Xu, Y.-X.; Xie, X.-L.; Wang, D.-F.; Aguilar-Gómez, D.; Liu, G.-J.; Li, X.; Esmailizadeh, A.; Rezaei, V.; Kantanen, J. Whole-genome sequence analysis unveils different origins of European and Asiatic mouflon and domestication-related genes in sheep. Commun. Biol. 2021, 4, 1–15. [Google Scholar] [CrossRef]
- Okarma, H.; Jędrzejewska, B.; Jędrzejewski, W.; Krasiński, Z.A.; Miłkowski, L. The roles of predation, snow cover, acorn crop, and man-related factors on ungulate mortality in Białowieża Primeval Forest, Poland. Acta Theriol. 1995, 40, 197–217. [Google Scholar] [CrossRef] [Green Version]
- Kays, R.; Curtis, A.; Kirchman, J.J. Rapid adaptive evolution of northeastern coyotes via hybridization with wolves. Biol. Lett. 2010, 6, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Barlow, A.; Cahill, J.A.; Hartmann, S.; Theunert, C.; Xenikoudakis, G.; Fortes, G.G.; Paijmans, J.L.; Rabeder, G.; Frischauf, C.; Grandal-d’Anglade, A. Partial genomic survival of cave bears in living brown bears. Nat. Ecol. Evol. 2018, 2, 1563–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Racimo, F.; Sankararaman, S.; Nielsen, R.; Huerta-Sánchez, E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015, 16, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.-S.; Wang, S.; Li, Y.; Jhala, Y.; Thakur, M.; Otecko, N.O.; Si, J.-F.; Chen, H.-M.; Shapiro, B.; Nielsen, R. Ancient hybridization with an unknown population facilitated high-altitude adaptation of canids. Mol. Biol. Evol. 2020, 37, 2616–2629. [Google Scholar] [CrossRef]
- Ferreira, M.S.; Jones, M.R.; Callahan, C.M.; Farelo, L.; Tolesa, Z.; Suchentrunk, F.; Boursot, P.; Mills, L.S.; Alves, P.C.; Good, J.M. The legacy of recurrent introgression during the radiation of hares. Syst. Biol. 2021, 70, 593–607. [Google Scholar] [CrossRef]
- Sankararaman, S.; Mallick, S.; Patterson, N.; Reich, D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 2016, 26, 1241–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumer, M.; Rosenthal, G.G.; Andolfatto, P. How common is homoploid hybrid speciation? Evolution 2014, 68, 1553–1560. [Google Scholar] [CrossRef]
- Macholán, M. Hybrid Zone, Mouse. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 588–591. [Google Scholar]
- Chang, S.W.; Oshida, T.; Endo, H.; Nguyen, S.; Dang, C.; Nguyen, D.; Jiang, X.; Li, Z.J.; Lin, L.K. Ancient hybridization and underestimated species diversity in Asian striped squirrels (genus Tamiops): Inference from paternal, maternal and biparental markers. J. Zool. 2011, 285, 128–138. [Google Scholar] [CrossRef]
- Detwiler, K.M. Mitochondrial DNA analyses of Cercopithecus monkeys reveal a localized hybrid origin for C. mitis doggetti in Gombe National Park, Tanzania. Int. J. Primatol. 2019, 40, 28–52. [Google Scholar] [CrossRef]
- Macholán, M.; Baird, S.J.; Dufková, P.; Munclinger, P.; Bímová, B.V.; Piálek, J. Assessing multilocus introgression patterns: A case study on the mouse X chromosome in central Europe. Evol. Int. J. Org. Evol. 2011, 65, 1428–1446. [Google Scholar] [CrossRef]
- Allen, W.L.; Stevens, M.; Higham, J.P. Character displacement of Cercopithecini primate visual signals. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komárek, J.; Komárková-Legnerová, J. Phenotype diversity of the cyanoprokaryotic genus Cylindrospermopsis (Nostocales). Czech Phycol. 2003, 3, 1–30. [Google Scholar]
- Ito, T.; Kawamoto, Y.; Hamada, Y.; Nishimura, T.D. Maxillary sinus variation in hybrid macaques: Implications for the genetic basis of craniofacial pneumatization. Biol. J. Linn. Soc. 2015, 115, 333–347. [Google Scholar] [CrossRef] [Green Version]
- Grant, P.R.; Grant, B.R. Phenotypic and genetic effects of hybridization in Darwin’s finches. Evolution 1994, 48, 297–316. [Google Scholar] [CrossRef]
- Gridley, T.; Elwen, S.H.; Harris, G.; Moore, D.; Hoelzel, A.; Lampen, F. Hybridization in bottlenose dolphins—A case study of Tursiops aduncus × T. truncatus hybrids and successful backcross hybridization events. PLoS ONE 2018, 13, e0201722. [Google Scholar] [CrossRef] [PubMed]
- Cserkész, T.; Kiss, C.; Barkaszi, Z.; Görföl, T.; Zagorodniuk, I.; Sramkó, G.; Csorba, G. Intra-and interspecific morphological variation in sympatric and allopatric populations of Mustela putorius and M. eversmanii (Carnivora: Mustelidae) and detection of potential hybrids. Mammal Res. 2021, 66, 103–114. [Google Scholar] [CrossRef]
- Balcarcel, A.; Sánchez-Villagra, M.R.; Segura, V.; Evin, A. Singular patterns of skull shape and brain size change in the domestication of South American camelids. J. Mammal. 2021, 102, 220–235. [Google Scholar] [CrossRef]
- Frare, C.F.; Matocq, M.D.; Feldman, C.R.; White, A.M.; Manley, P.N.; Jermstad, K.D.; Hekkala, E.R. Landscape disturbance and sporadic hybridization complicate field identification of chipmunks. J. Wildl. Manag. 2017, 81, 248–258. [Google Scholar] [CrossRef]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar] [CrossRef]
- Barton, N.H.; Hewitt, G.M. Adaptation, speciation and hybrid zones. Nature 1989, 341, 497–503. [Google Scholar] [CrossRef]
- Harrison, R. Hybrid Zones and the Evolutionary Process; Oxford University Press: New York, NY, USA, 1993. [Google Scholar]
- Woodruff, D.S. Natural hybridization and hybrid zones. Syst. Biol. 1973, 22, 213–218. [Google Scholar] [CrossRef]
- Mauldin, M.R.; Haynie, M.L.; Hanson, J.D.; Baker, R.J.; Bradley, R.D. Multilocus characterization of a woodrat (genus Neotoma) hybrid zone. J. Hered. 2014, 105, 466–476. [Google Scholar] [CrossRef] [Green Version]
- Shurtliff, Q.R.; Murphy, P.J.; Matocq, M.D. Ecological segregation in a small mammal hybrid zone: Habitat-specific mating opportunities and selection against hybrids restrict gene flow on a fine spatial scale. Evolution 2014, 68, 729–742. [Google Scholar] [CrossRef]
- Ďureje, Ľ.; Macholán, M.; Baird, S.J.; Piálek, J. The mouse hybrid zone in Central Europe: From morphology to molecules. J. Vertebr. Biol. 2012, 61, 308–318. [Google Scholar] [CrossRef]
- Coyner, B.S.; Murphy, P.J.; Matocq, M.D. Hybridization and asymmetric introgression across a narrow zone of contact between Neotoma fuscipes and N. macrotis (Rodentia: Cricetidae). Biol. J. Linn. Soc. 2015, 115, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Jahner, J.P.; Parchman, T.L.; Matocq, M.D. Multigenerational backcrossing and introgression between two woodrat species at an abrupt ecological transition. Mol. Ecol. 2021, 30, 4245–4258. [Google Scholar] [CrossRef]
- Mauldin, M.R.; Haynie, M.L.; Vrla, S.C.; Bradley, R.D. Temporal evaluation of a woodrat (genus Neotoma) hybrid zone based on genotypic and georeferenced data. J. Mammal. 2021, 102, 541–557. [Google Scholar] [CrossRef]
- Brandler, O.; Kapustina, S.; Nikol’skii, A.; Kolesnikov, V.; Badmaev, B.; Adiya, Y. A study of hybridization between Marmota baibacina and M. sibirica in their secondary contact zone in Mongolian Altai. Front. Ecol. Evol. 2021, 9, 363. [Google Scholar] [CrossRef]
- Gravena, W.; Da Silva, V.M.; Da Silva, M.N.; Farias, I.P.; Hrbek, T. Living between rapids: Genetic structure and hybridization in botos (Cetacea: Iniidae: Inia spp.) of the Madeira River, Brazil. Biol. J. Linn. Soc. 2015, 114, 764–777. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.D.; Cowan, P.; Gruber, B.; MacDonald, A.J.; Holleley, C.E.; Sarre, S.D. Has the introduction of two subspecies generated dispersal barriers among invasive possums in New Zealand? Biol. Invasions 2021, 23, 3831–3845. [Google Scholar] [CrossRef]
- Eldridge, M.D.; Pearson, D.J.; Potter, S. Identification of a novel hybrid zone within the black-footed rock-wallaby (Petrogale lateralis) in Western Australia. Aust. J. Zool. 2021, 68, 98–107. [Google Scholar] [CrossRef]
- Kinoshita, E.; Abramov, A.V.; Soloviev, V.A.; Saveljev, A.P.; Nishita, Y.; Kaneko, Y.; Masuda, R. Hybridization between the European and Asian badgers (Meles, Carnivora) in the Volga-Kama region, revealed by analyses of maternally, paternally and biparentally inherited genes. Mamm. Biol. 2019, 94, 140–148. [Google Scholar] [CrossRef]
- Baird, A.B.; Braun, J.K.; Engstrom, M.D.; Holbert, A.C.; Huerta, M.G.; Lim, B.K.; Mares, M.A.; Patton, J.C.; Bickham, J.W. Nuclear and mtDNA phylogenetic analyses clarify the evolutionary history of two species of native Hawaiian bats and the taxonomy of Lasiurini (Mammalia: Chiroptera). PLoS ONE 2017, 12, e0186085. [Google Scholar] [CrossRef]
- Lorenzini, R.; Fanelli, R.; Grifoni, G.; Scholl, F.; Fico, R. Wolf-dog crossbreeding: “Smelling” a hybrid may not be easy. Mamm. Biol. 2014, 79, 149–156. [Google Scholar] [CrossRef]
- Thompson, C.W.; Pfau, R.; Choate, J.R.; Genoways, H.H.; Finck, E.J. Identification and characterization of the contact zone between short-tailed shrews (Blarina) in Iowa and Missouri. Can. J. Zool. 2011, 89, 278–288. [Google Scholar] [CrossRef] [Green Version]
- Andriollo, T.; Ashrafi, S.; Arlettaz, R.; Ruedi, M. Porous barriers? Assessment of gene flow within and among sympatric long-eared bat species. Ecol. Evol. 2018, 8, 12841–12854. [Google Scholar] [CrossRef]
- Arbogast, B.S.; Schumacher, K.I.; Kerhoulas, N.J.; Bidlack, A.L.; Cook, J.A.; Kenagy, G. Genetic data reveal a cryptic species of New World flying squirrel: Glaucomys oregonensis. J. Mammal. 2017, 98, 1027–1041. [Google Scholar] [CrossRef] [Green Version]
- Yannic, G.; Statham, M.J.; Denoyelle, L.; Szor, G.; Qulaut, G.Q.; Sacks, B.N.; Lecomte, N. Investigating the ancestry of putative hybrids: Are Arctic fox and red fox hybridizing? Polar Biol. 2017, 40, 2055–2062. [Google Scholar] [CrossRef]
- Mengoni, C.; Mucci, N.; Randi, E. Genetic diversity and no evidences of recent hybridization in the endemic Italian hare (Lepus corsicanus). Conserv. Genet. 2015, 16, 477–489. [Google Scholar] [CrossRef]
- Prevosti, F.J.; Ramírez, M.A.; Schiaffini, M.; Martin, F.; Udrizar Sauthier, D.E.; Carrera, M.; Sillero-Zubiri, C.; Pardiñas, U.F. Extinctions in near time: New radiocarbon dates point to a very recent disappearance of the South American fox Dusicyon avus (Carnivora: Canidae). Biol. J. Linn. Soc. 2015, 116, 704–720. [Google Scholar] [CrossRef] [Green Version]
- Bell, K.C.; Van Gunst, J.; Teglas, M.B.; Hsueh, J.; Matocq, M.D. Lost in a sagebrush sea: Comparative genetic assessment of an isolated montane population of Tamias amoenus. J. Mammal. 2021, 102, 173–187. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, H.-Q.; Tu, X.-L.; Ji, C.-M.; Gou, X.; Esmailizadeh, A.; Wang, S.; Wang, M.-S.; Wang, M.-C.; Li, X.-L. Genomes reveal selective sweeps in kiang and donkey for high-altitude adaptation. Zool. Res. 2021, 42, 450. [Google Scholar] [CrossRef]
- Fabbri, E.; Velli, E.; D’Amico, F.; Galaverni, M.; Mastrogiuseppe, L.; Mattucci, F.; Caniglia, R. From predation to management: Monitoring wolf distribution and understanding depredation patterns from attacks on livestock. Hystrix Ital. J. Mammal. 2018, 29, 101–110. [Google Scholar]
- Inoue, T.; Murakami, T.; Abramov, A.V.; Masuda, R. Mitochondrial DNA control region variations in the sable Martes zibellina of Hokkaido Island and the Eurasian continent, compared with the Japanese marten M. melampus. Mammal Study 2010, 35, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, C.L.; Andersen, L.W.; Stronen, A.V. Forensic DNA analyses suggest illegal trade of canid skins. Mammal Res. 2016, 61, 423–426. [Google Scholar] [CrossRef]
- Eckert, I.; Suchentrunk, F.; Markov, G.; Hartl, G.B. Genetic diversity and integrity of German wildcat (Felis silvestris) populations as revealed by microsatellites, allozymes, and mitochondrial DNA sequences. Mamm. Biol. 2010, 75, 160–174. [Google Scholar] [CrossRef]
- Leite, J.V.; Álvares, F.; Velo-Antón, G.; Brito, J.C.; Godinho, R. Differentiation of North African foxes and population genetic dynamics in the desert—Insights into the evolutionary history of two sister taxa, Vulpes rueppellii and Vulpes vulpes. Org. Divers. Evol. 2015, 15, 731–745. [Google Scholar] [CrossRef]
- Sierra, A.B.A.; Castillo, E.R.; Labaroni, C.; Barrandeguy, M.E.; Martí, D.A.; Ojeda, R.; Lanzone, C. Genetic studies in the recently divergent Eligmodontia puerulus and E. moreni (Rodentia, Cricetidae, Sigmodontinae) from Puna and Monte deserts of South America. Mamm. Biol. 2017, 87, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Lawson, L.P.; Castruita, J.A.S.; Haile, J.S.; Vernesi, C.; Rovero, F.; Lorenzen, E.D. Unraveling elephant-shrews: Phylogenetic relationships and unexpected introgression among giant sengis. Mol. Phylogenet. Evol. 2021, 154, 107001. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.E.; Fenton, M.B.; Carstens, B.C.; Simmons, N.B. Comment on “Population genetics reveal Myotis keenii (Keen’s myotis) and Myotis evotis (long-eared myotis) to be a single species”. Can. J. Zool. 2021, 99, 415–422. [Google Scholar] [CrossRef]
- Nagata, J.; Yasuda, M.; Yamashiro, A. Genetic analysis of a newly established deer population expanding in the Sasebo area in Nagasaki Prefecture, Japan reveals no evidence of genetic disturbance by Formosan sika deer. Mamm. Study 2021, 46, 1–13. [Google Scholar] [CrossRef]
- Sarver, B.A.; Herrera, N.D.; Sneddon, D.; Hunter, S.S.; Settles, M.L.; Kronenberg, Z.; Demboski, J.R.; Good, J.M.; Sullivan, J. Diversification, introgression, and rampant cytonuclear discordance in rocky mountains chipmunks (sciuridae: Tamias). Syst. Biol. 2021, 70, 908–921. [Google Scholar] [CrossRef]
- Korablev, M.P.; Korablev, N.P.; Korablev, P.N. Genetic diversity and population structure of the grey wolf (Canis lupus Linnaeus, 1758) and evidence of wolf× dog hybridisation in the centre of European Russia. Mamm. Biol. 2021, 101, 91–104. [Google Scholar] [CrossRef]
- van Wyk, A.M.; Dalton, D.L.; Hoban, S.; Bruford, M.W.; Russo, I.R.M.; Birss, C.; Grobler, P.; van Vuuren, B.J.; Kotzé, A. Quantitative evaluation of hybridization and the impact on biodiversity conservation. Ecol. Evol. 2017, 7, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Furman, A.; Coraman, E.; Çelik, Y.E.; Postawa, T.; Bachanek, J.; Ruedi, M. Cytonuclear discordance and the species status of Myotis myotis and Myotis blythii (Chiroptera). Zool. Scr. 2014, 43, 549–561. [Google Scholar] [CrossRef]
- Furman, A.; Çelik, Y.E.; Çoraman, E.; Bilgin, R. Reproductive isolation and morphological discrimination of Myotis myotis macrocephalicus and M. blythii sl (Chiroptera: Vespertilionidae) in Turkey. Acta Chiropterol. 2020, 22, 21–28. [Google Scholar] [CrossRef]
- Burgarella, C.; Barnaud, A.; Kane, N.A.; Jankowski, F.; Scarcelli, N.; Billot, C.; Vigouroux, Y.; Berthouly-Salazar, C. Adaptive introgression: An untapped evolutionary mechanism for crop adaptation. Front. Plant Sci. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A.; Balding, D.J. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 2004, 13, 969–980. [Google Scholar] [CrossRef]
- Excoffier, L.; Hofer, T.; Foll, M. Detecting loci under selection in a hierarchically structured population. Heredity 2009, 103, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Browett, S.S.; O’Meara, D.B.; McDevitt, A.D. Genetic tools in the management of invasive mammals: Recent trends and future perspectives. Mamm. Rev. 2020, 50, 200–210. [Google Scholar] [CrossRef]
- Mattucci, F.; Galaverni, M.; Lyons, L.A.; Alves, P.C.; Randi, E.; Velli, E.; Pagani, L.; Caniglia, R. Genomic approaches to identify hybrids and estimate admixture times in European wildcat populations. Sci. Rep. 2019, 9, 1–15. [Google Scholar]



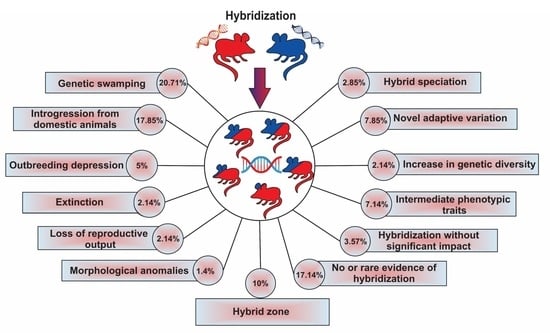
| Results | Impacts | Number of Papers | Percentage | Description | |
|---|---|---|---|---|---|
| 1 | Genetic swamping | Considered as negative | 29 | 20.71 | Genetic integrity of a species involved in hybridization being threatened by introgression from another species |
| 2 | Introgression from a domesticated lineage | Considered as negative | 25 | 17.85 | Genetic integrity of wild species being threatened by introgression from a domesticated lineage |
| 3 | Extinction due to extreme genetic swamping | Negative | 3 | 2.14 | Complete loss of genetic material of one of the species involved in hybridization |
| 4 | Outbreeding depression | Negative | 7 | 5.0 | Reduction or loss of specific adaptations and overall fitness |
| 5 | Morphological anomalies | Negative | 2 | 1.4 | Morphological anomalies with negative effects on fitness |
| 6 | Loss of reproductive output | Negative | 3 | 2.14 | Decrease in growth rate of parental species because of wasted reproductive effort |
| 7 | Increase in genetic diversity and reduction of inbreeding | Positive | 3 | 2.14 | Increase in genetic diversity via low rates of introgression, without any evidence of outbreeding depression; reduction of inbreeding levels |
| 8 | Gaining novel adaptive variation | Positive | 11 | 7.85 | Transferring of adaptive variants through hybridization |
| 9 | Hybrid speciation | Positive | 4 | 2.85 | Creation of a new species via hybridization |
| 10 | Intermediate phenotypic traits | Neutral or unknown | 10 | 7.14 | Intermediate morphological characteristics of hybrid individuals relative to the parental species |
| 11 | Hybrid zone | Neutral or unknown | 14 | 10.0 | Geographically restricted zones where genetically distinct species meet and mate |
| 12 | Hybridization without significant impacts | Neutral or unknown | 5 | 3.57 | Evidence of hybridization without substantial changes in the gene pools of each species |
| 13 | No or rare evidence of hybridization | Neutral or unknown | 24 | 17.14 | Hybridization is rare and does not result in introgression |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adavoudi, R.; Pilot, M. Consequences of Hybridization in Mammals: A Systematic Review. Genes 2022, 13, 50. https://doi.org/10.3390/genes13010050
Adavoudi R, Pilot M. Consequences of Hybridization in Mammals: A Systematic Review. Genes. 2022; 13(1):50. https://doi.org/10.3390/genes13010050
Chicago/Turabian StyleAdavoudi, Roya, and Małgorzata Pilot. 2022. "Consequences of Hybridization in Mammals: A Systematic Review" Genes 13, no. 1: 50. https://doi.org/10.3390/genes13010050