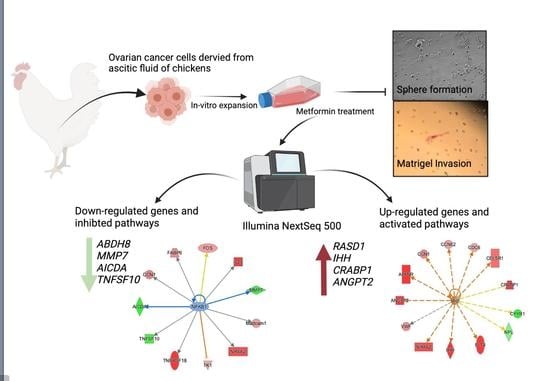
Metformin Affects the Transcriptomic Profile of Chicken Ovarian Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal, Isolation of COVCAR Cells
2.2. Primary Ovarian Cancer Cell Culture
2.3. Effect of Metformin on COVCAR Cell Sphere Formation
2.4. Effect of Metformin on Matrigel Invasion of COVCAR Cells
2.5. Effect of Metformin on COVCAR Cell Transcriptome
2.6. Data Visualization
3. Results
3.1. Effect of Metformin on Sphere Formation and Matrigel Invasion
3.2. Transcriptomic Analysis of Metformin-Treated COVCAR Cells
3.3. Identifying the Pathways Affected by Metformin Treatment Using Ingenuity Pathway Analysis (IPA)
3.3.1. AMPK Signaling in Ovarian Cancer Cells
3.3.2. Epithelial Mesenchymal Transition
3.3.3. Angiogenesis
3.3.4. Regulation of Apoptosis, DNA Damage Repair, and Cell-Cycle Progression
3.3.5. Causal Network Analysis to Identify Master Regulators Associated with Ovarian Cancer
3.3.6. Identification of Gene Networks Associated with Diseases and Cell Function
3.3.7. Identification of Biomarkers Using Bioprofiler in IPA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia, C.; Yao, A.; Camacho, F.; Balkrishnan, R.; Cantrell, L.A. A SEER-Medicare analysis of the impact of metformin on overall survival in ovarian cancer. Gynecol. Oncol. 2017, 146, 346–350. [Google Scholar] [CrossRef]
- Bailey, C.J. Metformin: Historical Overview. Diabetologia 2017, 60, 1566–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannucci, E.; Harlan, D.M.; Archer, M.C.; Bergenstal, R.M.; Gapstur, S.M.; Habel, L.A.; Pollak, M.; Regensteiner, J.G.; Yee, D. Diabetes and Cancer: A Consensus Report. CA Cancer J. Clin. 2010, 60, 207–221. [Google Scholar] [CrossRef] [Green Version]
- De Santi, M.; Baldelli, G.; Diotallevi, A.; Galluzzi, L.; Schiavano, G.F.; Brandi, G. Metformin prevents cell tumorigenesis through autophagy-related cell death. Sci. Rep. 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.B.; Lei, K.J.; Liu, J.P.; Jia, Y.M. Continuous use of metformin can improve survival in type 2 diabetic patients with ovarian cancer. Medicine 2017, 96, e7605. [Google Scholar] [CrossRef]
- Bodmer, M.; Becker, C.; Meier, C.; Jick, S.S.; Meier, C.R. Use of metformin and the risk of ovarian cancer: A case–control analysis. Gynecol. Oncol. 2011, 123, 200–204. [Google Scholar] [CrossRef]
- Brown, J.R.; Chan, D.K.; Shank, J.J.; Griffith, K.A.; Fan, H.; Szulawski, R.; Yang, K.; Reynolds, R.K.; Johnston, C.; McLean, K.; et al. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 2020, 5, e133247. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.L.; McCormick, A.; McEwen, K.A.; Park, S.; Karrison, T.; Yamada, S.D.; Pannain, S.; Lengyel, E. Relationship of Type II Diabetes and Metformin Use to Ovarian Cancer Progression, Survival, and Chemosensitivity. Obstet. Gynecol. 2012, 119, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Liu, B.; Wang, H.; Zhang, T.; Yang, L. Association of metformin use with ovarian cancer incidence and prognosis: A systematic review and meta-analysis. Int. J. Gynecol. Cancer 2019, 29, 140–146. [Google Scholar] [CrossRef]
- Tseng, C.H. Metformin reduces ovarian cancer risk in Taiwanese women with type 2 diabetes mellitus. Diabetes/Metab. Res. Rev. 2015, 31, 619–626. [Google Scholar] [CrossRef]
- Erices, R.; Bravo, M.L.; Gonzalez, P.; Oliva, B.; Racordon, D.; Garrido, M.; Ibañez, C.; Kato, S.; Brañes, J.; Pizarro, J.; et al. Metformin, at concentrations corresponding to the treatment of diabetes, potentiates the cytotoxic effects of carboplatin in cultures of ovarian cancer cells. Reprod. Sci. 2013, 20, 1433–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Zhao, N.; Li, D.; Zou, G.; Chen, Y. Metformin improves the sensitivity of ovarian cancer cells to chemotherapeutic agents. Oncol. Lett. 2019, 18, 2404–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shank, J.J.; Yang, K.; Ghannam, J.; Cabrera, L.; Johnston, C.J.; Reynolds, R.K.; Buckanovich, R.J. Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecol. Oncol. 2012, 127, 390–397. [Google Scholar] [CrossRef] [Green Version]
- Rattan, R.; Graham, R.P.; Maguire, J.L.; Giri, S.; Shridhar, V. Metformin suppresses ovarian cancer growth and metastasis with enhancement of cisplatin cytotoxicity in Vivo. Neoplasia 2011, 13, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderhyden, B.C.; Shaw, T.J.; Ethier, J.F. Animal models of ovarian cancer. Reprod. Biol. Endocrinol. 2003, 1, 67. [Google Scholar] [CrossRef] [Green Version]
- Mendes, N.; Dias Carvalho, P.; Martins, F.; Mendonça, S.; Malheiro, A.R.; Ribeiro, A.; Carvalho, J.; Velho, S. Animal Models to Study Cancer and Its Microenvironment. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Campbell, J.G. Some Unusual Gonadal Tumours of the Fowl. Br. J. Cancer 1951, 5, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, T.N. Ovarian Tumors of the Hen. Technical Report; 1987; Volume 73. Available online: https://ehp.niehs.nih.gov/doi/abs/10.1289/ehp.877335 (accessed on 1 October 2021).
- Barnes, M.N.; Berry, W.D.; Straughn, J.J.; Kirby, T.O.; Leath, C.A.; Huh, W.K.; Grizzle, W.E.; Partridge, E.E. A Pilot Study of Ovarian Cancer Chemoprevention Using Medroxyprogesterone Acetate in an Avian Model of Spontaneous Ovarian Carcinogenesis. Gynecol. Oncol. 2002, 87, 57–63. [Google Scholar] [CrossRef]
- Ansenberger, K.; Zhuge, Y.; Lagman, J.A.J.; Richards, C.; Barua, A.; Bahr, J.M.; Hales, D.B. E-cadherin expression in ovarian cancer in the laying hen, Gallus domesticus, compared to human ovarian cancer. Gynecol. Oncol. 2009, 113, 362–369. [Google Scholar] [CrossRef] [Green Version]
- Baker, H.; Fathalla, M.F. Veterinary Pathology; Technical Report; Campbell: Camden, NJ, USA, 1969; Volume 5, p. 272. [Google Scholar]
- Hakim, A.A.; Barry, C.P.; Barnes, H.J.; Anderson, K.E.; Petitte, J.; Whitaker, R.; Lancaster, J.M.; Wenham, R.M.; Carver, D.K.; Turbov, J.; et al. Ovarian Adenocarcinomas in the Laying Hen and Women Share Similar Alterations in p53, ras, and HER-2/neu. Cancer Prev. Res. 2009, 2, 114–121. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, A.; Hadley, J.A.; Hendricks, G.L.; Elkin, R.G.; Cooper, T.; Ramachandran, R. Characterization of Ascites-Derived Ovarian Tumor Cells from Spontaneously Occurring Ovarian Tumors of the Chicken: Evidence for E-Cadherin Upregulation. PLoS ONE 2013, 8, e57582. [Google Scholar] [CrossRef]
- Tiwari, A.; Hadley, J.A.; Ramachandran, R. Characterization of ascites-derived aldehyde dehydrogenase–positive ovarian cancer stem cells isolated from Leghorn chickens. Poult. Sci. 2020, 99, 2203–2214. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Stavnes, H.T.; Nymoen, D.A.; Falkenthal, T.E.; Kræn, J.; Tropé, C.G.; Davidson, B. APOA1 mRNA expression in ovarian serous carcinoma effusions is a marker of longer survival. Am. J. Clin. Pathol. 2014, 142, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Chan, C.H.; Chen, K.; Guan, X.; Lin, H.K.; Tong, Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 2012, 31, 1546–1557. [Google Scholar] [CrossRef]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the sirt1 deacetylase. Science 2004, 303, 5666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.S. SIRT1: Tumor promoter or tumor suppressor? Med. Hypotheses 2006, 67, 341–344. [Google Scholar] [CrossRef]
- Wang, F.; Nguyen, M.; Qin, F.X.F.; Tong, Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007, 6, 505–514. [Google Scholar] [CrossRef]
- Dang, J.H.; Jin, Z.J.; Liu, X.J.; Hu, D.; Wang, J.; Luo, Y.; Li, L.L. Metformin in combination with cisplatin inhibits cell viability and induces apoptosis of human ovarian cancer cells by inactivating ERK 1/2. Oncol. Lett. 2017, 14, 7557–7564. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.J.; Li, Y.Q.; Chen, Q.Y.; Xiao, S.J.; Zeng, S.E. Up-regulating of RASD1 and Apoptosis of DU-145 Human Prostate Cancer Cells Induced by Formononetin in Vitro. Asian Pac. J. Cancer Prev. 2014, 15, 2835–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Duan, Y.; Bei, C.; Chen, J. Calycosin Induces Apoptosis by Upregulation of RASD1 in Human Breast Cancer Cells MCF-7. Horm. Metab. Res. 2013, 45, 593–598. [Google Scholar] [CrossRef]
- Spinicelli, S.; Nocentini, G.; Ronchetti, S.; Krausz, L.T.; Bianchini, R.; Riccardi, C. GITR interacts with the pro-apoptotic protein Siva and induces apoptosis. Cell Death Differ. 2002, 9, 1382–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Paisie, T.K.; Chen, S.; Concannon, P. UBASH3A Regulates the Synthesis and Dynamics of TCR–CD3 Complexes. J. Immunol. 2019, 203, 2827–2836. [Google Scholar] [CrossRef]
- Collingwood, T.S.; Smirnova, E.V.; Bogush, M.; Carpino, N.; Annan, R.S.; Tsygankov, A.Y. T-cell Ubiquitin Ligand Affects Cell Death through a Functional Interaction with Apoptosis-inducing Factor, a Key Factor of Caspase-independent Apoptosis. J. Biol. Chem. 2007, 282, 30920–30928. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, E.D. An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs 1995, 154, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kan, T.; Wang, W.; Ip, P.P.; Zhou, S.; Wong, A.S.; Wang, X.; Yang, M. Single-cell EMT-related transcriptional analysis revealed intra-cluster heterogeneity of tumor cell clusters in epithelial ovarian cancer ascites. Oncogene 2020, 39, 4227–4240. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yang, Z.; Jin, P.; Yang, X.; Li, X.; Wei, X.; Wang, Y.; Long, S.; Zhang, T.; Chen, G.; et al. Metformin Suppresses Tumor Progression by Inactivating Stromal Fibroblasts in Ovarian Cancer. Mol. Cancer Ther. 2018, 17, 1291–1302. [Google Scholar] [CrossRef] [Green Version]
- Esparza-López, J.; Alvarado-Muñoz, J.F.; Escobar-Arriaga, E.; Ulloa-Aguirre, A.; de Jesús Ibarra-Sánchez, M. Metformin reverses mesenchymal phenotype of primary breast cancer cells through STAT3/NF-κB pathways. BMC Cancer 2019, 19, 728. [Google Scholar] [CrossRef] [PubMed]
- Phoenix, K.N.; Vumbaca, F.; Claffey, K.P. Therapeutic metformin/AMPK activation promotes the angiogenic phenotype in the ERα negative MDA-MB-435 breast cancer model. Breast Cancer Res. Treat. 2009, 113, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.J.; Hayward, R.; Viros, A.; Marais, R. Metformin Accelerates the Growth of BRAF V600E -Driven Melanoma by Upregulating VEGF-A. Cancer Discov. 2012, 2, 344–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunckhorst, M.K.; Xu, Y.; Lu, R.; Yu, Q. Angiopoietins promote ovarian cancer progression by establishing a procancer microenvironment. Am. J. Pathol. 2014, 184, 2285–2296. [Google Scholar] [CrossRef] [Green Version]
- Milam, K.E.; Parikh, S.M. The angiopoietin-Tie2 signaling axis in the vascular leakage of systemic inflammation. Tissue Barriers 2015, 3, e957508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorli, S.C.; Le Gonidec, S.; Knibiehler, B.; Audigier, Y. Apelin is a potent activator of tumour neoangiogenesis. Oncogene 2007, 26, 7692–7699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kälin, R.E.; Kretz, M.P.; Meyer, A.M.; Kispert, A.; Heppner, F.L.; Brändli, A.W. Paracrine and autocrine mechanisms of apelin signaling govern embryonic and tumor angiogenesis. Dev. Biol. 2007, 305, 599–614. [Google Scholar] [CrossRef] [Green Version]
- Neelakantan, D.; Dogra, S.; Devapatla, B.; Jaiprasart, P.; Mukashyaka, M.C.; Janknecht, R.; Dwivedi, S.K.D.; Bhattacharya, R.; Husain, S.; Ding, K.; et al. Multifunctional APJ Pathway Promotes Ovarian Cancer Progression and Metastasis. Mol. Cancer Res. 2019, 17, 1378–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Zeng, S.; Zheng, G.; Chen, D.; Li, P.; Yang, M.; Luo, K.; Yin, J.; Gu, Y.; Zhang, Z.; et al. FOXO3a-driven miRNA signatures suppresses VEGF-A/NRP1 signaling and breast cancer metastasis. Oncogene 2021, 40, 777–790. [Google Scholar] [CrossRef]
- Nelson, L.E.; Valentine, R.J.; Cacicedo, J.M.; Gauthier, M.S.; Ido, Y.; Ruderman, N.B. A novel inverse relationship between metformin-triggered AMPK-SIRT1 signaling and p53 protein abundance in high glucose-exposed HepG2 cells. Am. J. Physiol. Cell Physiol. 2012, 303, C4–C13. [Google Scholar] [CrossRef]
- Cerezo, M.; Tichet, M.; Abbe, P.; Ohanna, M.; Lehraiki, A.; Rouaud, F.; Allegra, M.; Giacchero, D.; Bahadoran, P.; Bertolotto, C.; et al. Metformin Blocks Melanoma Invasion and Metastasis Development in AMPK/p53-Dependent Manner. Mol. Cancer Ther. 2013, 12, 1605–1615. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar] [CrossRef]
- Rogoff, H.A.; Pickering, M.T.; Debatis, M.E.; Jones, S.; Kowalik, T.F. E2F1 Induces Phosphorylation of p53 That Is Coincident with p53 Accumulation and Apoptosis. Mol. Cell. Biol. 2002, 22, 5308–5318. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.; Kovacs, M.; Chellappan, S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: Role of acetylation. Cancer Res. 2010, 70, 4931–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, L.; Zhang, Y.; Wang, W.; Song, E.; Fan, Y.; Wei, B. E2F1: A promising regulator in ovarian carcinoma. Tumor Biol. 2016, 37, 2823–2831. [Google Scholar] [CrossRef]
- Timmers, C.; Sharma, N.; Opavsky, R.; Maiti, B.; Wu, L.; Wu, J.; Orringer, D.; Trikha, P.; Saavedra, H.I.; Leone, G. E2f1, E2f2, and E2f3 Control E2F Target Expression and Cellular Proliferation via a p53-Dependent Negative Feedback Loop. Mol. Cell. Biol. 2007, 27, 65–78. [Google Scholar] [CrossRef]
- Barsotti, A.M.; Prives, C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 2009, 28, 4295–4305. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Chang, T.; Wang, Y.; Liu, Y.; Li, W.; Li, M.; Fan, H.Y. YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PLoS ONE 2014, 9, e91770. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, Q.; Li, Y.; Wang, G.; Gao, S.; Zhang, X.; Yan, X.; Zhang, X.; Xie, J.; Wang, Y.; et al. Metformin targets a YAP1-TEAD4 complex via AMPKα to regulate CCNE1/2 in bladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 376. [Google Scholar] [CrossRef] [PubMed]








| Gene Name | Log2FC | p-Value | FDR | Function |
|---|---|---|---|---|
| ABHD8 | −6.5 | 6.12 | 7.60 | Influences methylation of 12CpG and increased EOC risk |
| MMP7 | −5.4 | 6.07 | 0.00 | An enzyme involved in extracellular matrix lysis |
| APOC3 | −5.3 | 1.55 | 6.00 | Involved in the metabolism of triglyceride (TG)-rich lipoproteins (TRLs) |
| AICDA | −4.8 | 3.93 | 2.47 | Involved in somatic hypermutation, gene conversion, and class-switch recombination of immunoglobulin genes in B-lymphocytes |
| GDPD2 | −4.5 | 2.28 | 3.80 | Plays a role in the remodeling of the actin cytoskeleton |
| APOA1 | −4.4 | 5.28 | 6.70 | Attaches to cell membranes and promotes the efflux of cholesterol and phospholipids from the cytoplasm |
| CYYR1 | −4.2 | 8.24 | 4.01 | Not annotated in the Gallus gallus genome |
| GSTA4 | −3.9 | 1.30 | 1.20 | Associated with hepatocellular carcinoma |
| S1PR3 | −3.7 | 1.96 | 3.50 | A G-protein-coupled receptor that contributes to the regulation of angiogenesis and vascular endothelial cell function |
| Gene Name | Log2FC | p-Value | FDR | Function |
|---|---|---|---|---|
| MNR2 | 5.5 | 1.85 | 3.40 | Regulation of transcription, sequence specific-DNA binding |
| SLC5A5 | 5.5 | 2.47 | 1.85 | A member of the sodium-glucose co-transporter family. Responsible for the uptake of iodine in thyroid and lactating breast tissue |
| TMEM82 | 4.7 | 2.37 | 8.00 | Transmembrane protein. Prognostic marker in Renal cancer (favorable) |
| IHH | 4.6 | 4.15 | 2.54 | Activation of the hedgehog pathway, implicated in the development of various cancers, prostate cancer, pancreatic cancer, gastrointestinal malignancies, and ovarian cancers |
| IL19 | 4.5 | 3.32 | 2.19 | Mitogenic and chemotactic for endothelial cells and can induce their angiogenic potential |
| UNC45B | 4.2 | 5.45 | 3.04 | Encodes a myosin-specific chaperone that, together with the general heat-shock protein HSP90, is involved in myosin assembly |
| CHRNB3 | 4.2 | 1.01 | 4.61 | Not annotated in the Gallus gallus genome |
| CRABP-I | 4.1 | 6.50 | 3.39 | CRABP1 is assumed to play an important role in retinoic acid-mediated differentiation and proliferation processes |
| RSAD2 | 4 | 3.29 | 1.10 | Induced by interferon, known to abolish amino acid and mitochondrial metabolism |
| SLC6A17 | 4 | 3.80 | 0.00 | A member of the SLC6 family of transporters, responsible for the presynaptic uptake of most neurotransmitters |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gopalan, L.; Sebastian, A.; Praul, C.A.; Albert, I.; Ramachandran, R. Metformin Affects the Transcriptomic Profile of Chicken Ovarian Cancer Cells. Genes 2022, 13, 30. https://doi.org/10.3390/genes13010030
Gopalan L, Sebastian A, Praul CA, Albert I, Ramachandran R. Metformin Affects the Transcriptomic Profile of Chicken Ovarian Cancer Cells. Genes. 2022; 13(1):30. https://doi.org/10.3390/genes13010030
Chicago/Turabian StyleGopalan, Lalitha, Aswathy Sebastian, Craig A. Praul, Istvan Albert, and Ramesh Ramachandran. 2022. "Metformin Affects the Transcriptomic Profile of Chicken Ovarian Cancer Cells" Genes 13, no. 1: 30. https://doi.org/10.3390/genes13010030