Investigating Markers of the NLRP3 Inflammasome Pathway in Alzheimer’s Disease: A Human Post-Mortem Study
Abstract
:1. Introduction
2. Results
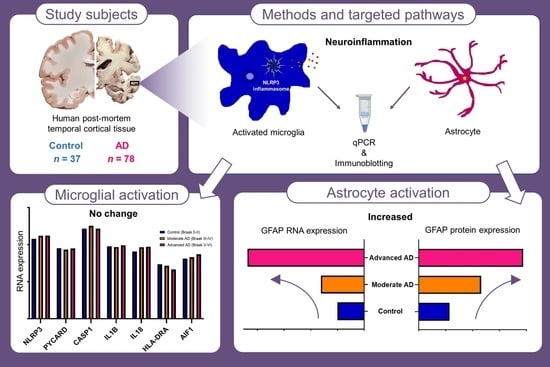
2.1. RNA and Protein Expression of GFAP Was Increased in Advanced AD
2.2. RNA Expression of AIF1 and HLA-DRA Was Unchanged in AD
2.3. RNA and Protein Expression of NLRP3 Inflammasome Was Unchanged in AD
3. Discussion
4. Materials and Methods
4.1. Source of Human Brain Tissue and Research Ethics Committee Approval
4.2. Quantitative Real-Time PCR
4.2.1. RNA Isolation and cDNA Synthesis
4.2.2. Selection of Candidate Reference Genes and PCR Primer Design
4.2.3. Reverse Transcriptase Semi-Quantitative Real-time PCR (RT-qPCR)
4.3. Protein Analyses: Capillary Electrophoresis Immunoblotting
4.3.1. Protein Extraction and Purification
4.3.2. Protein Quantification
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reitz, C.; Brayne, C.; Mayeux, R. Epidemiology of Alzheimer Disease. Nat. Rev. Neurol. 2011, 7, 137–152. [Google Scholar] [CrossRef]
- Alzheimers Dement. 2021 Alzheimer’s Disease Facts and Figures. Alzheimers Dement. 2021, 17, 327–406. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. A Century of Alzheimer’s Disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M. Neuroinflammation in Alzheimer’s Disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Bazan, N.G.; Halabi, A.; Ertel, M.; Petasis, N.A. Chapter 34—Neuroinflammation. In Basic Neurochemistry; Academic Press: Cambridge, MA, USA, 2012; pp. 610–620. [Google Scholar] [CrossRef]
- Morales, I.; Jiménez, J.M.; Mancilla, M.; Maccioni, R.B. Tau Oligomers and Fibrils Induce Activation of Microglial Cells. J. Alzheimers Dis. 2013, 37, 849–856. [Google Scholar] [CrossRef]
- Welikovitch, L.A.; Do Carmo, S.; Maglóczky, Z.; Malcolm, J.C.; Lőke, J.; Klein, W.L.; Freund, T.; Cuello, A.C. Early Intraneuronal Amyloid Triggers Neuron-Derived Inflammatory Signaling in APP Transgenic Rats and Human Brain. Proc. Natl. Acad. Sci. USA 2020, 117, 6844–6854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.; Beja-Glasser, V.F.; Nfonoyim, B.M.; Frouin, A.; Li, S.; Ramakrishnan, S.; Merry, K.M.; Shi, Q.; Rosenthal, A.; Barres, B.A. Complement and Microglia Mediate Early Synapse Loss in Alzheimer Mouse Models. Science 2016, 352, 712–716. [Google Scholar] [CrossRef] [Green Version]
- Glass, C.K.; Saijo, K.; Winner, B.; Marchetto, M.C.; Gage, F.H. Mechanisms Underlying Inflammation in Neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Zhu, Y.; Lin, N.; Zhang, J.; Ye, Q.; Huang, H.; Chen, X. Microglial Phagocytosis Induced by Fibrillar β-Amyloid Is Attenuated by Oligomeric β-Amyloid: Implications for Alzheimer’s Disease. Mol. Neurodegener. 2011, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kempuraj, D.; Thangavel, R.; Natteru, P.A.; Selvakumar, G.P.; Saeed, D.; Zahoor, H.; Zaheer, S.; Iyer, S.S.; Zaheer, A. Neuroinflammation Induces Neurodegeneration. J. Neurol. Neurosurg. Spine 2016, 1, 1003. [Google Scholar]
- Hornung, V.; Bauernfeind, F.; Halle, A.; Samstad, E.O.; Kono, H.; Rock, K.L.; Fitzgerald, K.A.; Latz, E. Silica Crystals and Aluminum Salts Activate the NALP3 Inflammasome through Phagosomal Destabilization. Nat. Immunol. 2008, 9, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, V.A.K.; Fitzgerald, K.A. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 2016, 165, 792–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoss, F.; Rodriguez-Alcazar, J.F.; Latz, E. Assembly and Regulation of ASC Specks. Cell. Mol. Life Sci. 2017, 74, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Dixit, V.M. Mechanisms and Functions of Inflammasomes. Cell 2014, 157, 1013–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneka, M.T.; McManus, R.M.; Latz, E. Inflammasome Signalling in Brain Function and Neurodegenerative Disease. Nat. Rev. Neurosci. 2018, 19, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Kummer, M.P.; Stutz, A.; Delekate, A.; Schwartz, S.; Vieira-Saecker, A.; Griep, A.; Axt, D.; Remus, A.; Tzeng, T.-C. NLRP3 Is Activated in Alzheimer’s Disease and Contributes to Pathology in APP/PS1 Mice. Nature 2013, 493, 674–678. [Google Scholar] [CrossRef]
- Venegas, C.; Kumar, S.; Franklin, B.S.; Dierkes, T.; Brinkschulte, R.; Tejera, D.; Vieira-Saecker, A.; Schwartz, S.; Santarelli, F.; Kummer, M.P. Microglia-Derived ASC Specks Cross-Seed Amyloid-β in Alzheimer’s Disease. Nature 2017, 552, 355–361. [Google Scholar] [CrossRef]
- Ising, C.; Venegas, C.; Zhang, S.; Scheiblich, H.; Schmidt, S.v.; Vieira-Saecker, A.; Schwartz, S.; Albasset, S.; McManus, R.M.; Tejera, D. NLRP3 Inflammasome Activation Drives Tau Pathology. Nature 2019, 575, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Halle, A.; Hornung, V.; Petzold, G.C.; Stewart, C.R.; Monks, B.G.; Reinheckel, T.; Fitzgerald, K.A.; Latz, E.; Moore, K.J.; Golenbock, D.T. The NALP3 Inflammasome Is Involved in the Innate Immune Response to Amyloid-β. Nat. Immunol. 2008, 9, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Stancu, I.-C.; Cremers, N.; Vanrusselt, H.; Couturier, J.; Vanoosthuyse, A.; Kessels, S.; Lodder, C.; Brône, B.; Huaux, F.; Octave, J.-N. Aggregated Tau Activates NLRP3–ASC Inflammasome Exacerbating Exogenously Seeded and Non-Exogenously Seeded Tau Pathology in Vivo. Acta Neuropathol. 2019, 137, 599–617. [Google Scholar] [CrossRef] [Green Version]
- Cribbs, D.H.; Berchtold, N.C.; Perreau, V.; Coleman, P.D.; Rogers, J.; Tenner, A.J.; Cotman, C.W. Extensive Innate Immune Gene Activation Accompanies Brain Aging, Increasing Vulnerability to Cognitive Decline and Neurodegeneration: A Microarray Study. J. Neuroinflammation 2012, 9, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chan, C. IPAF Inflammasome Is Involved in Interleukin-1β Production from Astrocytes, Induced by Palmitate; Implications for Alzheimer’s Disease. Neurobiol. Aging 2014, 35, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Braak, H.; Thal, D.R.; Ghebremedhin, E.; del Tredici, K. Stages of the Pathologic Process in Alzheimer Disease: Age Categories from 1 to 100 Years. J. Neuropathol. Exp. Neurol. 2011, 70, 960–969. [Google Scholar] [CrossRef]
- Sajdel-Sulkowska, E.M.; Majocha, R.E.; Salim, M.; Zain, S.B.; Marotta, C.A. The Postmortem Alzheimer Brain Is a Source of Structurally and Functionally Intact Astrocytic Messenger RNA. J. Neurosci. Methods 1988, 23, 173–179. [Google Scholar] [CrossRef]
- Coulson, D.T.R.; Beyer, N.; Quinn, J.G.; Brockbank, S.; Hellemans, J.; Irvine, G.B.; Ravid, R.; Johnston, J.A. BACE1 MRNA Expression in Alzheimer’s Disease Postmortem Brain Tissue. J. Alzheimers Dis. 2010, 22, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.E.; Ince, P.G.; Lace, G.; Forster, G.; Shaw, P.J.; Matthews, F.; Savva, G.; Brayne, C.; Wharton, S.B. Astrocyte Phenotype in Relation to Alzheimer-Type Pathology in the Ageing Brain. Neurobiol. Aging 2010, 31, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Perez-Nievas, B.G.; Serrano-Pozo, A. Deciphering the Astrocyte Reaction in Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porchet, R.; Probst, A.; Bouras, C.; Dráberová, E.; Dráber, P.; Riederer, B.M. Analysis of Gial Acidic Fibrillary Protein in the Human Entorhinal Cortex during Aging and in Alzheimer’s Disease. Proteomics 2003, 3, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Oeckl, P.; Halbgebauer, S.; Anderl-Straub, S.; Steinacker, P.; Huss, A.M.; Neugebauer, H.; von Arnim, C.A.F.; Diehl-Schmid, J.; Grimmer, T.; Kornhuber, J. Glial Fibrillary Acidic Protein in Serum Is Increased in Alzheimer’s Disease and Correlates with Cognitive Impairment. J. Alzheimers Dis. 2019, 67, 481–488. [Google Scholar] [CrossRef]
- Pereira, J.B.; Janelidze, S.; Smith, R.; Mattsson-Carlgren, N.; Palmqvist, S.; Teunissen, C.E.; Zetterberg, H.; Stomrud, E.; Ashton, N.J.; Blennow, K. Plasma GFAP Is an Early Marker of Amyloid-β but Not Tau Pathology in Alzheimer’s Disease. Brain 2021, awab223. [Google Scholar] [CrossRef]
- Cicognola, C.; Janelidze, S.; Hertze, J.; Zetterberg, H.; Blennow, K.; Mattsson-Carlgren, N.; Hansson, O. Plasma Glial Fibrillary Acidic Protein Detects Alzheimer Pathology and Predicts Future Conversion to Alzheimer Dementia in Patients with Mild Cognitive Impairment. Alzheimers Res. Ther. 2021, 13, 1–9. [Google Scholar] [CrossRef]
- Hopperton, K.E.; Mohammad, D.; Trépanier, M.O.; Giuliano, V.; Bazinet, R.P. Markers of Microglia in Post-Mortem Brain Samples from Patients with Alzheimer’s Disease: A Systematic Review. Mol. Psychiatry 2018, 23, 177–198. [Google Scholar] [CrossRef]
- Sanchez-Mejias, E.; Navarro, V.; Jimenez, S.; Sanchez-Mico, M.; Sanchez-Varo, R.; Nuñez-Diaz, C.; Trujillo-Estrada, L.; Davila, J.; Vizuete, M.; Gutierrez, A.; et al. Soluble phospho-tau from Alzheimer’s disease hippocampus drives microglial degeneration. Acta Neuropathol. 2016, 132, 897–916. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, H.; Mackay, M.; Palmer, K.; Tothová, K.; Katsur, M.; Platt, B.; Koss, D.J. Synaptic Loss, ER Stress and Neuro-Inflammation Emerge Late in the Lateral Temporal Cortex and Associate with Progressive Tau Pathology in Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 3258–3272. [Google Scholar] [CrossRef]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.B.; Li, G. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A. New Tools for Studying Microglia in the Mouse and Human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ismael, S.; Nasoohi, S.; Sakata, K.; Liao, F.-F.; McDonald, M.P.; Ishrat, T. Thioredoxin-Interacting Protein (TXNIP) Associated NLRP3 Inflammasome Activation in Human Alzheimer’s Disease Brain. J. Alzheimers Dis. 2019, 68, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Ojala, J.; Alafuzoff, I.; Herukka, S.-K.; van Groen, T.; Tanila, H.; Pirttilä, T. Expression of Interleukin-18 Is Increased in the Brains of Alzheimer’s Disease Patients. Neurobiol. Aging. 2009, 30, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Friedman, B.; Srinivasan, K.; David, H. Human Alzheimer’s Disease Microglial Activation Is Not Fully Captured by Existing Mouse Models. In GLIA; Wiley: Hoboken, NJ, USA, 2019; Volume 67, pp. E632–E633. [Google Scholar]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E. Identification of a Unique TGF-β–Dependent Molecular and Functional Signature in Microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransohoff, R.M. How Neuroinflammation Contributes to Neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Slanzi, A.; Iannoto, G.; Rossi, B.; Zenaro, E.; Constantin, G. In Vitro Models of Neurodegenerative Diseases. Front. Cell. Dev. Biol. 2020, 8, 328. [Google Scholar] [CrossRef]
- Xu, X.; Nehorai, A.; Dougherty, J.D. Cell type-specific analysis of human brain transcriptome data to predict alterations in cellular composition. Syst. Biomed. 2013, 1, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on MRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preece, P.; Cairns, N.J. Quantifying MRNA in Postmortem Human Brain: Influence of Gender, Age at Death, Postmortem Interval, Brain PH, Agonal State and Inter-Lobe MRNA Variance. Mol. Brain Res. 2003, 118, 60–71. [Google Scholar] [CrossRef]
- Koppelkamm, A.; Vennemann, B.; Lutz-Bonengel, S.; Fracasso, T.; Vennemann, M. RNA Integrity in Post-Mortem Samples: Influencing Parameters and Implications on RT-QPCR Assays. Int. J. Legal. Med. 2011, 125, 573–580. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Allawi, H.T.; SantaLucia, J. Thermodynamics and NMR of Internal G.T Mismatches in DNA. Biochemistry 1997, 36, 10581–10594. [Google Scholar] [CrossRef] [Green Version]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Section 3.5, Purifying, Detecting, and Characterizing Proteins. In Molecular Cell Biology; WH Freeman: New York, NY, USA, 2000. [Google Scholar]
- Harris, V.M. Protein Detection by Simple WesternTM Analysis. Methods Mol. Biol. 2015, 1312, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





| Study | Sample Size | Brain Region | Technique | Markers of NLRP3 Inflammasome | ||
|---|---|---|---|---|---|---|
| NLRP3 | PYCARD/ASC | CASP1/Caspase-1 | ||||
| Heneka et al. [17] | AD, n = 12 NC, n = 8 | FC HC | WB | NR | NR | ↑ in AD |
| Cribbs et all. [22] | AD, n = 26 NC, n = 33 | EC | Microarray | ↑ in NC | ||
| HC | ||||||
| SFG | ||||||
| PCG | ||||||
| SFG | qPCR | no change | no change | |||
| Liu et al. [23] | AD, n = 5 NC, n = 5 | FC FC | qPCR WB | no change no change | ↑ in AD ↑ in AD | NR |
| Li et al. [38] | AD, n = 6–7 NC, n = 6–7 | FC | WB | no change | ↑ in AD | ↑ in AD |
| Present study | AD, n = 78 NC, n = 37 | TC | qPCR | no change | no change | no change |
| Age of Death (Years) | Post-Mortem Interval (Hours) | APOE Genotype | ||
|---|---|---|---|---|
| ε4 Non-Carrier | ε4 Carrier | |||
| Control group | ||||
| All (n = 37) | 82.6 ± 11.5 | 81.2 ± 36.9 | 32 | 5 |
| Male (n = 19) | 84.5 ± 9.4 | 85.7 ± 41.4 | 17 | 2 |
| Female (n = 18) | 80.7 ± 13.3 | 76.5 ± 31.8 | 15 | 3 |
| Moderate AD group | ||||
| All (n = 39) | 85.5 ± 6.3 | 79.4 ± 38.4 | 21 | 18 |
| Male (n = 20) | 84.0 ± 6.5 | 81.2 ± 37.0 | 9 | 11 |
| Female (n = 19) | 87.1 ± 6.0 | 77.5 ± 40.6 | 12 | 7 |
| Advanced AD group | ||||
| All (n = 39) | 78.1 ± 8.8 * | 86.2 ± 43.0 | 10 | 29 |
| Male (n = 20) | 73.3 ± 4.7 * | 82.7 ± 48.9 | 4 | 16 |
| Female (n = 19) | 83.0 ± 9.3 | 89.6 ± 37.1 | 6 | 13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, H.; Harte, M. Investigating Markers of the NLRP3 Inflammasome Pathway in Alzheimer’s Disease: A Human Post-Mortem Study. Genes 2021, 12, 1753. https://doi.org/10.3390/genes12111753
Tang H, Harte M. Investigating Markers of the NLRP3 Inflammasome Pathway in Alzheimer’s Disease: A Human Post-Mortem Study. Genes. 2021; 12(11):1753. https://doi.org/10.3390/genes12111753
Chicago/Turabian StyleTang, Hao, and Michael Harte. 2021. "Investigating Markers of the NLRP3 Inflammasome Pathway in Alzheimer’s Disease: A Human Post-Mortem Study" Genes 12, no. 11: 1753. https://doi.org/10.3390/genes12111753