Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs
Abstract
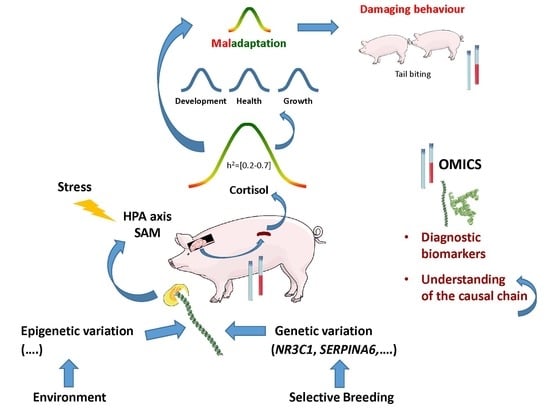
:1. Introduction
2. The Importance of Omics Technologies for Ethical Pig Production
3. Biomarkers for Stress Resilience in Farm Animals
4. Genetics of Stress Response
4.1. Quantitative and Molecular Genetics
4.2. Functional Genomics
5. Omics to Study Tail Biting
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Merks, J.W.M.; Mathur, P.K.; Knol, E.F. New phenotypes for new breeding goals in pigs. Animal 2012, 6, 535–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friggens, N.C.; Blanc, F.; Berry, D.P.; Puillet, L. Review: Deciphering animal robustness. A synthesis to facilitate its use in livestock breeding and management. Animal 2017, 11, 2237–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheffer, M.; Bolhuis, J.E.; Borsboom, D.; Buchman, T.G.; Gijzel, S.M.W.; Goulson, D.; Kammenga, J.E.; Kemp, B.; van de Leemput, I.A.; Levin, S.; et al. Quantifying resilience of humans and other animals. Proc. Natl. Acad. Sci. USA 2018, 115, 11883–11890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mormede, P.; Terenina, E. Molecular genetics of the adrenocortical axis and breeding for robustness. Domest. Anim. Endocrinol. 2012, 43, 116–131. [Google Scholar] [CrossRef]
- Wolf, C.; Linden, D.E.J. Biological pathways to adaptability—Interactions between genome, epigenome, nervous system and environment for adaptive behavior. Genes Brain Behav. 2012, 11, 3–28. [Google Scholar] [CrossRef]
- Rauw, W.M.; Kanis, E.; Noordhuizen-Stassen, E.N.; Grommers, F.J. Undesirable side effects of selection for high production efficiency in farm animals: A review. Livest. Prod. Sci. 1998, 56, 15–33. [Google Scholar] [CrossRef]
- Breuer, K.; Sutcliffe, M.E.M.; Mercer, J.T.; Rance, K.A.; O’Connell, N.E.; Sneddon, I.A.; Edwards, S.A. Heritability of clinical tail-biting and its relation to performance traits. Livest. Prod. Sci. 2005, 93, 87–94. [Google Scholar] [CrossRef]
- Nakov, D.; Hristov, S.; Stankovic, B.; Pol, F.; Dimitrov, I.; Ilieski, V.; Mormede, P.; Hervé, J.; Terenina, E.; Lieubeau, B.; et al. Methodologies for Assessing Disease Tolerance in Pigs. Front. Vet. Sci. 2018, 5, 329. [Google Scholar] [CrossRef]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2019, 9, 692. [Google Scholar] [CrossRef] [Green Version]
- Valros, A. Chapter 5—Tail biting. In Advances in Pig Welfare; Herd and Flock Welfare; Špinka, M., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 137–166. ISBN 978-0-08-101012-9. [Google Scholar]
- D’Eath, R.B.; Arnott, G.; Turner, S.P.; Jensen, T.; Lahrmann, H.P.; Busch, M.E.; Niemi, J.K.; Lawrence, A.B.; Sandøe, P. Injurious tail biting in pigs: How can it be controlled in existing systems without tail docking? Animal 2014, 8, 1479–1497. [Google Scholar] [CrossRef] [Green Version]
- Jensen, P.; Buitenhuis, B.; Kjaer, J.; Zanella, A.; Mormède, P.; Pizzari, T. Genetics and genomics of animal behaviour and welfare—Challenges and possibilities. Appl. Anim. Behav. Sci. 2008, 112, 383–403. [Google Scholar] [CrossRef]
- Turner, S.P.; Camerlink, I.; Baxter, E.M.; D’Eath, R.B.; Desire, S.; Roehe, R. 14—Breeding for pig welfare: Opportunities and challenges. In Advances in Pig Welfare; Herd and Flock Welfare; Špinka, M., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 399–414. ISBN 978-0-08-101012-9. [Google Scholar]
- Kanis, E.; De Greef, K.H.; Hiemstra, A.; van Arendonk, J.A.M. Breeding for societally important traits in pigs. J. Anim. Sci. 2005, 83, 948–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, N.R.; Parker, R.M.A.; Mendl, M.; Edwards, S.A.; Main, D.C.J. Prevalence of risk factors for tail biting on commercial farms and intervention strategies. Vet. J. 2012, 194, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Grümpel, A.; Krieter, J.; Dippel, S. Reducing estimated tail biting risk in German weaner pigs using a management tool. Vet. J. 2019, 254, 105406. [Google Scholar] [CrossRef]
- Hettinga, K.; Zhang, L. Omics and Systems Biology: Integration of Production and Omics Data in Systems Biology. In Proteomics in Domestic Animals: From Farm to Systems Biology; de Almeida, A.M., Eckersall, D., Miller, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 463–485. ISBN 978-3-319-69682-9. [Google Scholar]
- van der Steen, H.A.M.; Prall, G.F.W.; Plastow, G.S. Application of genomics to the pork industry. J. Anim. Sci. 2005, 83, E1–E8. [Google Scholar] [CrossRef]
- Parreira, J.R.; de Sousa Araújo, S. Studying the Animal Transcriptome: State of the Art and Challenges in the Context of Animal and Veterinary Sciences. In Proteomics in Domestic Animals: From Farm to Systems Biology; de Almeida, A.M., Eckersall, D., Miller, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 421–446. ISBN 978-3-319-69682-9. [Google Scholar]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Stamps, J.; Groothuis, T.G.G. The development of animal personality: Relevance, concepts and perspectives. Biol. Rev. 2010, 85, 301–325. [Google Scholar] [CrossRef] [Green Version]
- LaFreniere, P.; MacDonald, K. A post-genomic view of behavioral development and adaptation to the environment. Dev. Rev. 2013, 33, 89–109. [Google Scholar] [CrossRef]
- Meaney, M.J.; Szyf, M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005, 28, 456–463. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Moreno-Moral, A.; Petretto, E. From integrative genomics to systems genetics in the rat to link genotypes to phenotypes. Model. Mech. 2016, 9, 1097–1110. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.; Schiffer, L.; Re, A.; Azhar, R.; Basunia, A.; Rodriguez, C.; Chan, T.; Chapman, P.; Davis, S.R.; Gomez-Cabrero, D.; et al. Software for the Integration of Multiomics Experiments in Bioconductor. Cancer Res. 2017, 77, e39–e42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17, S15. [Google Scholar] [CrossRef] [Green Version]
- Tini, G.; Marchetti, L.; Priami, C.; Scott-Boyer, M.-P. Multi-omics integration—A comparison of unsupervised clustering methodologies. Brief. Bioinform. 2019, 20, 1269–1279. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [Green Version]
- Montastier, E.; Villa-Vialaneix, N.; Caspar-Bauguil, S.; Hlavaty, P.; Tvrzicka, E.; Gonzalez, I.; Saris, W.H.M.; Langin, D.; Kunesova, M.; Viguerie, N. System model network for adipose tissue signatures related to weight changes in response to calorie restriction and subsequent weight maintenance. PLoS Comput. Biol. 2015, 11, e1004047. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis—A framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef]
- Peng, C.; Wang, J.; Asante, I.; Louie, S.; Jin, R.; Chatzi, L.; Casey, G.; Thomas, D.C.; Conti, D.V. A latent unknown clustering integrating multi-omics data (LUCID) with phenotypic traits. Bioinformatics 2020, 36, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Ferrer, C.; Ruiz-Arenas, C.; Beltran-Gomila, A.; González, J.R. MultiDataSet: An R package for encapsulating multiple data sets with application to omic data integration. BMC Bioinform. 2017, 18, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukunaga, T.; Iwasaki, W. Logicome Profiler: Exhaustive detection of statistically significant logic relationships from comparative omics data. PLoS ONE 2020, 15, e0232106. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Smith, J.A.; Zhou, X. Leveraging gene co-expression patterns to infer trait-relevant tissues in genome-wide association studies. PLOS Genet. 2020, 16, e1008734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aten, J.E.; Fuller, T.F.; Lusis, A.J.; Horvath, S. Using genetic markers to orient the edges in quantitative trait networks: The NEO software. BMC Syst. Biol. 2008, 2, 34. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Tenenhaus, A.; Philippe, C.; Guillemot, V.; Le Cao, K.-A.; Grill, J.; Frouin, V. Variable selection for generalized canonical correlation analysis. Biostatistics 2014, 15, 569–583. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef]
- Martinez-Miro, S.; Tecles, F.; Ramon, M.; Escribano, D.; Hernandez, F.; Madrid, J.; Orengo, J.; Martinez-Subiela, S.; Manteca, X.; Joaquin Ceron, J. Causes, consequences and biomarkers of stress in swine: An update. BMC Vet. Res. 2016, 12, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimsa, U.; Tuchscherer, M.; Kanitz, E. Psychosocial Stress and Immunity—What Can We Learn From Pig Studies? Front. Behav. Neurosci. 2018, 12, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marco-Ramell, A.; de Almeida, A.M.; Cristobal, S.; Rodrigues, P.; Roncada, P.; Bassols, A. Proteomics and the search for welfare and stress biomarkers in animal production in the one-health context. Mol. Biosyst. 2018, 12, 2024–2035. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.S. Computer Models of Stress, Allostasis, and Acute and Chronic Diseases. Ann. N. Y. Acad. Sci. 2008, 1148, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Mato, A.; Salgado, F.J.; López-Pedrouso, M.; Carrera, M.; Bravo, S.; Parrado, M.; Gallardo, J.M.; Zapata, C. Tackling proteome changes in the longissimus thoracis bovine muscle in response to pre-slaughter stress. J. Proteomics 2015, 122, 73–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mato, A.; Rodríguez-Vázquez, R.; López-Pedrouso, M.; Bravo, S.; Franco, D.; Zapata, C. The first evidence of global meat phosphoproteome changes in response to pre-slaughter stress. BMC Genom. 2019, 20, 590. [Google Scholar] [CrossRef] [Green Version]
- Fuente-Garcia, C.; Aldai, N.; Sentandreu, E.; Oliván, M.; García-Torres, S.; Franco, D.; Zapata, C.; Sentandreu, M.A. Search for proteomic biomarkers related to bovine pre-slaughter stress using liquid isoelectric focusing (OFFGEL) and mass spectrometry. J. Proteom. 2019, 198, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wijffels, G.; Yu, Y.; Nielsen, L.K.; Niemeyer, D.O.; Fisher, A.D.; Ferguson, D.M.; Schirra, H.J. Altered fatty acid metabolism in long duration road transport: An NMR-based metabonomics study in sheep. J. Proteome Res. 2011, 10, 1073–1087. [Google Scholar] [CrossRef]
- Hao, Y.; Feng, Y.; Yang, P.; Cui, Y.; Liu, J.; Yang, C.; Gu, X. Transcriptome analysis reveals that constant heat stress modifies the metabolism and structure of the porcine longissimus dorsi skeletal muscle. Mol. Genet. Genom. 2016, 291, 2101–2115. [Google Scholar] [CrossRef]
- Cui, Y.; Hao, Y.; Li, J.; Bao, W.; Li, G.; Gao, Y.; Gu, X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. Int. J. Mol. Sci. 2016, 17, 393. [Google Scholar] [CrossRef] [Green Version]
- Qu, H.; Ajuwon, K.M. Metabolomics of heat stress response in pig adipose tissue reveals alteration of phospholipid and fatty acid composition during heat stress1. J. Anim. Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sandercock, D.A.; Barnett, M.W.; Coe, J.E.; Downing, A.C.; Nirmal, A.J.; Di Giminiani, P.; Edwards, S.A.; Freeman, T.C. Transcriptomics Analysis of Porcine Caudal Dorsal Root Ganglia in Tail Amputated Pigs Shows Long-Term Effects on Many Pain-Associated Genes. Front. Vet. Sci. 2019, 6, 314. [Google Scholar] [CrossRef] [PubMed]
- Escribano, D.; Horvatić, A.; Contreras-Aguilar, M.D.; Guillemin, N.; Cerón, J.J.; Tecles, F.; Martinez-Miró, S.; Eckersall, P.D.; Manteca, X.; Mrljak, V. Changes in saliva proteins in two conditions of compromised welfare in pigs: An experimental induced stress by nose snaring and lameness. Res. Vet. Sci. 2019, 125, 227–234. [Google Scholar] [CrossRef]
- de Almeida, A.M.; Bendixen, E. Pig proteomics: A review of a species in the crossroad between biomedical and food sciences. J. Proteom. 2012, 75, 4296–4314. [Google Scholar] [CrossRef]
- Lamy, E.; Mau, M. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteom. 2012, 75, 4251–4258. [Google Scholar] [CrossRef]
- Redei, E.E. Molecular genetics of the stress-responsive adrenocortical axis. Ann. Med. 2008, 40, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Murani, E.; Reyer, H.; Ponsuksili, S.; Fritschka, S.; Wimmers, K. A substitution in the ligand binding domain of the porcine glucocorticoid receptor affects activity of the adrenal gland. PLoS ONE 2012, 7, e45518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauw, W.M.; Johnson, A.K.; Gomez-Raya, L.; Dekkers, J.C.M. A Hypothesis and Review of the Relationship between Selection for Improved Production Efficiency, Coping Behavior, and Domestication. Front. Genet. 2017, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Ruis, M.A.W.; te Brake, J.H.A.; Engel, B.; Buist, W.G.; Blokhuis, H.J.; Koolhaas, J.M. Adaptation to social isolation: Acute and long-term stress responses of growing gilts with different coping characteristics. Physiol. Behav. 2001, 73, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Smolinska, A.; Blanchet, L.; Buydens, L.M.C.; Wijmenga, S.S. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef]
- Mischak, H.; Allmaier, G.; Apweiler, R.; Attwood, T.; Baumann, M.; Benigni, A.; Bennett, S.E.; Bischoff, R.; Bongcam-Rudloff, E.; Capasso, G.; et al. Recommendations for Biomarker Identification and Qualification in Clinical Proteomics. Sci. Transl. Med. 2010, 2, 46ps42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Diorio, J.; Tannenbaum, B.; Caldji, C.; Francis, D.; Freedman, A.; Sharma, S.; Pearson, D.; Plotsky, P.M.; Meaney, M.J. Maternal Care, Hippocampal Glucocorticoid Receptors, and Hypothalamic-Pituitary-Adrenal Responses to Stress. Science 1997, 277, 1659–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, I.C.G.; Meaney, M.J.; Szyf, M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc. Natl. Acad. Sci. USA 2006, 103, 3480–3485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suderman, M.; McGowan, P.O.; Sasaki, A.; Huang, T.C.T.; Hallett, M.T.; Meaney, M.J.; Turecki, G.; Szyf, M. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc. Natl. Acad. Sci. USA 2012, 109, 17266–17272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Giménez, J.L.; Seco-Cervera, M.; Tollefsbol, T.O.; Romá-Mateo, C.; Peiró-Chova, L.; Lapunzina, P.; Pallardó, F.V. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2017, 54, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Ritsner, M.S.; Gottesman, I.I. Where Do We Stand in the Quest for Neuropsychiatric Biomarkers and Endophenotypes and What Next. In The Handbook of Neuropsychiatric Biomarkers, Endophenotypes and Genes: Neuropsychological Endophenotypes and Biomarkers; Ritsner, M.S., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 3–21. ISBN 978-1-4020-9464-4. [Google Scholar]
- Logue, M.W.; Miller, M.W.; Wolf, E.J.; Huber, B.R.; Morrison, F.G.; Zhou, Z.; Zheng, Y.; Smith, A.K.; Daskalakis, N.P.; Ratanatharathorn, A.; et al. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin. Epigenet. 2020, 12, 46. [Google Scholar] [CrossRef]
- Otten, W.; Kanitz, E.; Tuchscherer, M. The impact of pre-natal stress on offspring development in pigs. J. Agric. Sci. 2015, 153, 907–919. [Google Scholar] [CrossRef]
- Schachtschneider, K.M.; Welge, M.E.; Auvil, L.S.; Chaki, S.; Rund, L.A.; Madsen, O.; Elmore, M.R.P.; Johnson, R.W.; Groenen, M.A.M.; Schook, L.B. Altered Hippocampal Epigenetic Regulation Underlying Reduced Cognitive Development in Response to Early Life Environmental Insults. Genes 2020, 11, 162. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.J.; Kelly, G.; Sengupta, A.; Heydendael, W.; Nicholas, B.; Beltrami, S.; Luz, S.; Peixoto, L.; Abel, T.; Bhatnagar, S. MicroRNAs as biomarkers of resilience or vulnerability to stress. Neuroscience 2015, 305, 36–48. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, U.; Keck, M.E.; Buell, D.R. miRNAs and other non-coding RNAs in posttraumatic stress disorder: A systematic review of clinical and animal studies. J. Psychiatr. Res. 2015, 65, 1–8. [Google Scholar] [CrossRef]
- Lecchi, C.; Zamarian, V.; Gini, C.; Avanzini, C.; Polloni, A.; Rota Nodari, S.; Ceciliani, F. Salivary microRNAs are potential biomarkers for the accurate and precise identification of inflammatory response after tail docking and castration in piglets. J. Anim. Sci. 2020, 98. [Google Scholar] [CrossRef] [PubMed]
- Schöler, N.; Langer, C.; Döhner, H.; Buske, C.; Kuchenbauer, F. Serum microRNAs as a novel class of biomarkers: A comprehensive review of the literature. Exp. Hematol. 2010, 38, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Ai, S.; Shen, L.; Guo, J.; Feng, X.; Tang, B. DNA Methylation as a Biomarker for Neuropsychiatric Diseases. Int. J. Neurosci. 2012, 122, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, A.; Auta, J.; Davis, J.M.; Dong, E.; Gavin, D.P.; Grayson, D.R.; Sharma, R.P.; Smith, R.C.; Tueting, P.; Zhubi, A. Toward the Identification of Peripheral Epigenetic Biomarkers of Schizophrenia. J. Neurogenet. 2014, 28, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Ahanda, M.-L.E.; Zerjal, T.; Dhorne-Pollet, S.; Rau, A.; Cooksey, A.; Giuffra, E. Impact of the Genetic Background on the Composition of the Chicken Plasma MiRNome in Response to a Stress. PLoS ONE 2014, 9, e114598. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.; Sánchez-Molano, E.; Psifidi, A.; Donadeu, F.X.; Banos, G. Association of plasma microRNA expression with age, genetic background and functional traits in dairy cattle. Sci. Rep. 2018, 8, 12955. [Google Scholar] [CrossRef]
- Murani, E.; Trakooljul, N.; Hadlich, F.; Ponsuksili, S.; Wimmers, K. Transcriptome Responses to Dexamethasone Depending on Dose and Glucocorticoid Receptor Sensitivity in the Liver. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Bianco, E.; Nevado, B.; Ramos-Onsins, S.E.; Pérez-Enciso, M. A Deep Catalog of Autosomal Single Nucleotide Variation in the Pig. PLoS ONE 2015, 10, e0118867. [Google Scholar] [CrossRef] [Green Version]
- Weiler, U.; Claus, R.; Schnoebelen-Combes, S.; Louveau, I. Influence of age and genotype on endocrine parameters and growth performance: A comparative study in Wild boars, Meishan and Large White boars. Livest. Prod. Sci. 1998, 54, 21–31. [Google Scholar] [CrossRef]
- Künzl, C.; Sachser, N. The Behavioral Endocrinology of Domestication: A Comparison between the Domestic Guinea Pig (Cavia apereaf.porcellus) and Its Wild Ancestor, the Cavy (Cavia aperea). Horm. Behav. 1999, 35, 28–37. [Google Scholar] [CrossRef]
- Malmkvist, J.; Hansen, S.W. The Welfare of Farmed Mink (Mustela Vison) in Relation to Behavioural Selection: A Review. Available online: https://www.ingentaconnect.com/content/ufaw/aw/2001/00000010/00000001/art00004 (accessed on 11 May 2020).
- Albert, F.W.; Carlborg, Ö.; Plyusnina, I.; Besnier, F.; Hedwig, D.; Lautenschläger, S.; Lorenz, D.; McIntosh, J.; Neumann, C.; Richter, H.; et al. Genetic Architecture of Tameness in a Rat Model of Animal Domestication. Genetics 2009, 182, 541–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trut, L.; Oskina, I.; Kharlamova, A. Animal evolution during domestication: The domesticated fox as a model. Bioessays 2009, 31, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsaesser, F.; Pfaffl, M.W.; Meyer, H.H.D.; Serpek, B.; Sauerwein, H. Differences in the somatotropic axis, in blood cortisol, insulin and thyroid hormone concentrations between two pig genotypes with markedly divergent growth rates and the effects of growth hormone treatment. Anim. Sci. 2002, 74, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Foury, A.; Geverink, N.A.; Gil, M.; Gispert, M.; Hortós, M.; Furnols, M.F.I.; Carrion, D.; Blott, S.C.; Plastow, G.S.; Mormède, P. Stress neuroendocrine profiles in five pig breeding lines and the relationship with carcass composition. Animal 2007, 1, 973–982. [Google Scholar] [CrossRef] [Green Version]
- Colpoys, J.; Van Sambeek, D.; Bruns, C.; Johnson, A.; Dekkers, J.; Dunshea, F.; Gabler, N. Responsiveness of swine divergently selected for feed efficiency to exogenous adrenocorticotropic hormone and glucose challenges. Domest. Anim. Endocrinol. 2018, 68, 32–38. [Google Scholar] [CrossRef]
- Leenhouwers, J.I.; Knol, E.F.; de Groot, P.N.; Vos, H.; van der Lende, T. Fetal development in the pig in relation to genetic merit for piglet survival. J. Anim. Sci. 2002, 80, 1759–1770. [Google Scholar] [CrossRef]
- Leenhouwers, J.I.; Knol, E.F.; van der Lende, T. Differences in late prenatal development as an explanation for genetic differences in piglet survival. Livest. Prod. Sci. 2002, 78, 57–62. [Google Scholar] [CrossRef]
- Lebret, B.; Ecolan, P.; Bonhomme, N.; Meteau, K.; Prunier, A. Influence of production system in local and conventional pig breeds on stress indicators at slaughter, muscle and meat traits and pork eating quality. Animal 2015, 9, 1404–1413. [Google Scholar] [CrossRef] [Green Version]
- Devillers, N.; Le Dividich, J.; Prunier, A. Influence of colostrum intake on piglet survival and immunity. Animal 2011, 5, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Kadarmideen, H.N.; Janss, L.L.G. Population and systems genetics analyses of cortisol in pigs divergently selected for stress. Physiol. Genom. 2007, 29, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Larzul, C.; Terenina, E.; Foury, A.; Billon, Y.; Louveau, I.; Merlot, E.; Mormede, P. The cortisol response to ACTH in pigs, heritability and influence of corticosteroid-binding globulin. Animal 2015, 9, 1929–1934. [Google Scholar] [CrossRef] [Green Version]
- Désautés, C.; Bidanel, J.P.; Milan, D.; Iannuccelli, N.; Amigues, Y.; Bourgeois, F.; Caritez, J.C.; Renard, C.; Chevalet, C.; Mormède, P. Genetic linkage mapping of quantitative trait loci for behavioral and neuroendocrine stress response traits in pigs. J. Anim. Sci. 2002, 80, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.P.; Iannuccelli, N.; Basso, B.; Foury, A.; Billon, Y.; Gandemer, G.; Gilbert, H.; Mormède, P.; Bidanel, J.P.; Larzul, C.; et al. Microsatellite mapping of quantitative trait loci affecting meat quality, stress hormones and production traits in Duroc × Large White F2 pigs. Animal 2011, 5, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamura, T.; Onodera, W.; Tayama, T.; Kadowaki, H.; Kojima-Shibata, C.; Suzuki, E.; Uemoto, Y.; Mikawa, S.; Hayashi, T.; Awata, T.; et al. A genome-wide scan for quantitative trait loci affecting respiratory disease and immune capacity in Landrace pigs. Anim. Genet. 2012, 43, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Muráni, E.; Ponsuksili, S.; D’Eath, R.B.; Turner, S.P.; Kurt, E.; Evans, G.; Thölking, L.; Klont, R.; Foury, A.; Mormède, P.; et al. Association of HPA axis-related genetic variation with stress reactivity and aggressive behaviour in pigs. BMC Genet. 2010, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Ponsuksili, S.; Du, Y.; Murani, E.; Schwerin, M.; Wimmers, K. Elucidating molecular networks that either affect or respond to plasma cortisol concentration in target tissues of liver and muscle. Genetics 2012, 192, 1109–1122. [Google Scholar] [CrossRef] [Green Version]
- Hessing, M.J.; Hagelsø, A.M.; Schouten, W.G.; Wiepkema, P.R.; van Beek, J.A. Individual behavioral and physiological strategies in pigs. Physiol. Behav. 1994, 55, 39–46. [Google Scholar] [CrossRef]
- Zebunke, M.; Repsilber, D.; Nuernberg, G.; Wittenburg, D.; Puppe, B. The backtest in pigs revisited—An analysis of intra-situational behaviour. Appl. Anim. Behav. Sci. 2015, 169, 17–25. [Google Scholar] [CrossRef]
- Rohrer, G.A.; Brown-Brandl, T.; Rempel, L.A.; Schneider, J.F.; Holl, J. Genetic analysis of behavior traits in swine production. Livest. Sci. 2013, 157, 28–37. [Google Scholar] [CrossRef]
- Scheffler, K.; Stamer, E.; Traulsen, I.; Krieter, J. Genetic analysis of the individual pig behaviour in backtests and human approach tests. Appl. Anim. Behav. Sci. 2014, 160, 38–45. [Google Scholar] [CrossRef]
- Velie, B.D.; Maltecca, C.; Cassady, J.P. Genetic relationships among pig behavior, growth, backfat, and loin muscle area. J. Anim. Sci. 2009, 87, 2767–2773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponsuksili, S.; Zebunke, M.; Murani, E.; Trakooljul, N.; Krieter, J.; Puppe, B.; Schwerin, M.; Wimmers, K. Integrated Genome-wide association and hypothalamus eQTL studies indicate a link between the circadian rhythm-related gene PER1 and coping behavior. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Putz, A.M.; Harding, J.C.S.; Dyck, M.K.; Fortin, F.; Plastow, G.S.; Dekkers, J.C.M.; Canada, P. Novel Resilience Phenotypes Using Feed Intake Data from a Natural Disease Challenge Model in Wean-to-Finish Pigs. Front. Genet. 2018, 9, 660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, A.J.; Keel, B.N.; Brown-Brandl, T.M.; Cassady, J.P.; Rohrer, G.A. Genome-wide association of changes in swine feeding behaviour due to heat stress. Genet. Sel. Evol. 2018, 50, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyer, H.; Ponsuksili, S.; Kanitz, E.; Pöhland, R.; Wimmers, K.; Murani, E. A Natural Mutation in Helix 5 of the Ligand Binding Domain of Glucocorticoid Receptor Enhances Receptor-Ligand Interaction. PLoS ONE 2016, 11, e0164628. [Google Scholar] [CrossRef]
- Muráni, E.; Ponsuksili, S.; Jaeger, A.; Görres, A.; Tuchscherer, A.; Wimmers, K. A naturally hypersensitive glucocorticoid receptor elicits a compensatory reduction of hypothalamus–pituitary–adrenal axis activity early in ontogeny. Open Biol. 2016, 6, 150193. [Google Scholar] [CrossRef]
- Ousova, O.; Guyonnet-Duperat, V.; Iannuccelli, N.; Bidanel, J.-P.; Milan, D.; Genêt, C.; Llamas, B.; Yerle, M.; Gellin, J.; Chardon, P.; et al. Corticosteroid Binding Globulin: A New Target for Cortisol-Driven Obesity. Mol. Endocrinol. 2004, 18, 1687–1696. [Google Scholar] [CrossRef] [Green Version]
- Esteve, A.; Ojeda, A.; Huang, L.S.; Folch, J.M.; Pérez-Enciso, M. Nucleotide variability of the porcine SERPINA6 gene and the origin of a putative causal mutation associated with meat quality. Anim. Genet. 2011, 42, 235–241. [Google Scholar] [CrossRef]
- Guyonnet-Dupérat, V.; Geverink, N.; Plastow, G.S.; Evans, G.; Ousova, O.; Croisetière, C.; Foury, A.; Richard, E.; Mormède, P.; Moisan, M.-P. Functional Implication of an Arg307Gly Substitution in Corticosteroid-Binding Globulin, a Candidate Gene for a Quantitative Trait Locus Associated with Cortisol Variability and Obesity in Pig. Genetics 2006, 173, 2143–2149. [Google Scholar] [CrossRef] [Green Version]
- Görres, A.; Ponsuksili, S.; Wimmers, K.; Muráni, E. Analysis of non-synonymous SNPs of the porcine SERPINA6 gene as potential causal variants for a QTL affecting plasma cortisol levels on SSC7. Anim. Genet. 2015, 46, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; van Binsbergen, R.; Brøndum, R.F.; Liao, X.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Nicolae, D.L.; Gamazon, E.; Zhang, W.; Duan, S.; Dolan, M.E.; Cox, N.J. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010, 6, e1000888. [Google Scholar] [CrossRef]
- Albert, F.W.; Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015, 16, 197–212. [Google Scholar] [CrossRef]
- Emilsson, V.; Thorleifsson, G.; Zhang, B.; Leonardson, A.S.; Zink, F.; Zhu, J.; Carlson, S.; Helgason, A.; Walters, G.B.; Gunnarsdottir, S.; et al. Genetics of gene expression and its effect on disease. Nature 2008, 452, 423–428. [Google Scholar] [CrossRef]
- Schadt, E.E. Novel integrative genomics strategies to identify genes for complex traits. Anim. Genet. 2006, 37, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.C.; Nap, J.-P. Genetical genomics: The added value from segregation. Trends Genet. 2001, 17, 388–391. [Google Scholar] [CrossRef] [Green Version]
- Gamazon, E.R.; Ziliak, D.; Im, H.K.; LaCroix, B.; Park, D.S.; Cox, N.J.; Huang, R.S. Genetic Architecture of MicroRNA Expression: Implications for the Transcriptome and Complex Traits. Am. J. Hum. Genet. 2012, 90, 1046–1063. [Google Scholar] [CrossRef] [Green Version]
- Murani, E.; Ponsuksili, S.; Reyer, H.; Wittenburg, D.; Wimmers, K. Expression variation of the porcine ADRB2 has a complex genetic background. Mol. Genet. Genom. 2013, 288, 615–625. [Google Scholar] [CrossRef]
- Jaeger, A.; Fritschka, S.; Ponsuksili, S.; Wimmers, K.; Muráni, E. Identification and Functional Characterization of Cis-Regulatory Elements Controlling Expression of the Porcine ADRB2 Gene. Int. J. Biol. Sci. 2015, 11, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffra, E.; Tuggle, C.K.; Consortium, T.F. Functional Annotation of Animal Genomes (FAANG): Current Achievements and Roadmap. Annu. Rev. Anim. Biosci. 2019, 17, 65–88. [Google Scholar] [CrossRef] [PubMed]
- Groß, C.; Derks, M.; Megens, H.-J.; Bosse, M.; Groenen, M.A.M.; Reinders, M.; de Ridder, D. pCADD: SNV prioritisation in Sus scrofa. Genet. Sel. Evol. 2020, 52, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, N.; Martini, J.W.R.; Zhang, Z.; Yuan, X.; Zhang, H.; Simianer, H.; Li, J. Incorporating Gene Annotation into Genomic Prediction of Complex Phenotypes. Genetics 2017, 207, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Weighting sequence variants based on their annotation increases the power of genome-wide association studies in dairy cattle. Genet. Sel. Evol. 2019, 51, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowles, C.R.; Hirschhorn, J.N.; Altshuler, D.; Lander, E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002, 32, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Maroilley, T.; Lemonnier, G.; Lecardonnel, J.; Esquerré, D.; Ramayo-Caldas, Y.; Mercat, M.J.; Rogel-Gaillard, C.; Estellé, J. Deciphering the genetic regulation of peripheral blood transcriptome in pigs through expression genome-wide association study and allele-specific expression analysis. BMC Genom. 2017, 18, 967. [Google Scholar] [CrossRef]
- Crowley, J.J.; Zhabotynsky, V.; Sun, W.; Huang, S.; Pakatci, I.K.; Kim, Y.; Wang, J.R.; Morgan, A.P.; Calaway, J.D.; Aylor, D.L.; et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 2015, 47, 353–360. [Google Scholar] [CrossRef]
- Brunberg, E.I.; Rodenburg, T.B.; Rydhmer, L.; Kjaer, J.B.; Jensen, P.; Keeling, L.J. Omnivores Going Astray: A Review and New Synthesis of Abnormal Behavior in Pigs and Laying Hens. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Nalon, E.; De Briyne, N. Efforts to Ban the Routine Tail Docking of Pigs and to Give Pigs Enrichment Materials via EU Law: Where Do We Stand a Quarter of a Century on? Animals 2019, 9, 132. [Google Scholar] [CrossRef] [Green Version]
- EFSA. Scientific report on the risks associated with tail biting in pigs and possible means to reduce the need for tail docking considering the different housing and husbandry systems. EFSA J. 2007, 5, 611. [Google Scholar] [CrossRef]
- Sinisalo, A.; Niemi, J.K.; Heinonen, M.; Valros, A. Tail biting and production performance in fattening pigs. Livest. Sci. 2012, 143, 220–225. [Google Scholar] [CrossRef]
- Breuer, K.; Sutcliffe, M.E.M.; Mercer, J.T.; Rance, K.A.; Beattie, V.E.; Sneddon, I.A.; Edwards, S.A. The effect of breed on the development of adverse social behaviours in pigs. Appl. Anim. Behav. Sci. 2003, 84, 59–74. [Google Scholar] [CrossRef]
- Moinard, C.; Mendl, M.; Nicol, C.J.; Green, L.E. A case control study of on-farm risk factors for tail biting in pigs. Appl. Anim. Behav. Sci. 2003, 81, 333–355. [Google Scholar] [CrossRef]
- Brunberg, E. Tail Biting and Feather Pecking. Available online: https://pub.epsilon.slu.se/8319/ (accessed on 17 June 2020).
- Wurtz, K.E.; Siegford, J.M.; Ernst, C.W.; Raney, N.E.; Bates, R.O.; Steibel, J.P. Genome-wide association analyses of lesion counts in group-housed pigs. Anim. Genet. 2018, 49, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.; Zanella, R.; Ventura, C.; Johansen, H.L.; Framstad, T.; Janczak, A.; Zanella, A.J.; Neibergs, H.L. Identification of chromosomal locations associated with tail biting and being a victim of tail-biting behaviour in the domestic pig (Sus scrofa domesticus). J. Appl. Genet. 2012, 53, 449–456. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, C.; Zhang, Z.; Wang, J.; Wang, Y.; Li, X.; Gao, R.; Wang, Z.; Fang, Y.; Wang, J.; et al. Blood-based dynamic genomic signature for obsessive–compulsive disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018, 177, 709–716. [Google Scholar] [CrossRef]
- Zhang-James, Y.; Fernàndez-Castillo, N.; Hess, J.L.; Malki, K.; Glatt, S.J.; Cormand, B.; Faraone, S.V. An integrated analysis of genes and functional pathways for aggression in human and rodent models. Mol. Psychiatry 2019, 24, 1655–1667. [Google Scholar] [CrossRef]
- Fogel, B.L.; Wexler, E.; Wahnich, A.; Friedrich, T.; Vijayendran, C.; Gao, F.; Parikshak, N.; Konopka, G.; Geschwind, D.H. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 2012, 21, 4171–4186. [Google Scholar] [CrossRef] [Green Version]
- Fernàndez-Castillo, N.; Gan, G.; van Donkelaar, M.M.J.; Vaht, M.; Weber, H.; Retz, W.; Meyer-Lindenberg, A.; Franke, B.; Harro, J.; Reif, A.; et al. RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior. Eur. Neuropsychopharmacol. 2020, 30, 44–55. [Google Scholar] [CrossRef]
- Brunberg, E.; Jensen, P.; Isaksson, A.; Keeling, L.J. Behavioural and Brain Gene Expression Profiling in Pigs during Tail Biting Outbreaks—Evidence of a Tail Biting Resistant Phenotype. PLoS ONE 2013, 8, e66513. [Google Scholar] [CrossRef] [PubMed]
- Brunberg, E.; Jensen, P.; Isaksson, A.; Keeling, L.J. Brain gene expression differences are associated with abnormal tail biting behavior in pigs: Gene expression and abnormal behavior in pigs. Genes Brain Behav. 2013, 12, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Lei, M.G.; Zhang, Y.B.; Wang, J.H.; Feng, X.T.; Xu, D.Q.; Gui, J.F.; Xiong, Y.Z. Characterization of the porcine differentially expressed PDK4 gene and association with meat quality. Mol. Biol. Rep. 2009, 36, 2003–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, T.; Dorr, N.P.; Takahashi, N.; McInnes, L.A.; Elder, G.A.; Buxbaum, J.D. Haploinsufficiency of Gtf2i, a gene deleted in Williams Syndrome, leads to increases in social interactions. Autism Res. 2011, 4, 28–39. [Google Scholar] [CrossRef]
- Hao, Y.; Liu, J.R.; Zhang, Y.; Yang, P.G.; Feng, Y.J.; Cui, Y.J.; Yang, C.H.; Gu, X.H. The microRNA expression profile in porcine skeletal muscle is changed by constant heat stress. Anim. Genet. 2016, 47, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Keltikangas-Jarvinen, L.; Puttonen, S.; Kivimaki, M.; Rontu, R.; Lehtimaki, T. Cloninger’s temperament dimensions and epidermal growth factor A61G polymorphism in Finnish adults. Genes Brain Behav. 2006, 5, 11–18. [Google Scholar] [CrossRef]
- Brunberg, E.; Jensen, P.; Isaksson, A.; Keeling, L. Feather pecking behavior in laying hens: Hypothalamic gene expression in birds performing and receiving pecks. Poult. Sci. 2011, 90, 1145–1152. [Google Scholar] [CrossRef]
- Štrac, D.Š.; Pivac, N.; Mück-Šeler, D. The serotonergic system and cognitive function. Transl. Neurosci. 2016, 7, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Pani, L.; Porcella, A.; Gessa, G.L. The role of stress in the pathophysiology of the dopaminergic system. Mol. Psychiatry 2000, 5, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Pezze, M.A.; Feldon, J. Mesolimbic dopaminergic pathways in fear conditioning. Prog. Neurobiol. 2004, 74, 301–320. [Google Scholar] [CrossRef]
- Ursinus, W.W.; Van Reenen, C.G.; Reimert, I.; Bolhuis, J.E. Tail biting in pigs: Blood serotonin and fearfulness as pieces of the puzzle? PLoS ONE 2014, 9, e107040. [Google Scholar] [CrossRef] [PubMed]
- Valros, A.; Palander, P.; Heinonen, M.; Munsterhjelm, C.; Brunberg, E.; Keeling, L.; Piepponen, P. Evidence for a link between tail biting and central monoamine metabolism in pigs (Sus scrofa domestica). Physiol. Behav. 2015, 143, 151–157. [Google Scholar] [CrossRef] [PubMed]
- de Haas, E.N.; van der Eijk, J.A.J. Where in the serotonergic system does it go wrong? Unravelling the route by which the serotonergic system affects feather pecking in chickens. Neurosci. Biobehav. Rev. 2018, 95, 170–188. [Google Scholar] [CrossRef] [PubMed]
- Kjaer, J.B.; Hjarvard, B.M.; Jensen, K.H.; Hansen-Møller, J.; Naesbye Larsen, O. Effects of haloperidol, a dopamine D2 receptor antagonist, on feather pecking behaviour in laying hens. Appl. Anim. Behav. Sci. 2004, 86, 77–91. [Google Scholar] [CrossRef]
- Flisikowski, K.; Schwarzenbacher, H.; Wysocki, M.; Weigend, S.; Preisinger, R.; Kjaer, J.B.; Fries, R. Variation in neighbouring genes of the dopaminergic and serotonergic systems affects feather pecking behaviour of laying hens. Anim. Genet. 2009, 40, 192–199. [Google Scholar] [CrossRef]
- Palander, P.A.; Heinonen, M.; Simpura, I.; Edwards, S.A.; Valros, A.E. Jejunal morphology and blood metabolites in tail biting, victim and control pigs. Animal 2013, 7, 1523–1531. [Google Scholar] [CrossRef] [Green Version]



| Name | Characteristics | Integration of Types of Omics | Type of Analysis | Reference and URL |
|---|---|---|---|---|
| Cytoscape | standalone software | Mainly protein–protein, protein–DNA and DNA–DNA interactions, but plug-ins (apps) are available for all types of omics | Provides tools to visualize complex molecular and genetic interaction networks, but also network analysis, enrichment analysis, ontology analysis and pathway analysis (e.g., KEGG) is possible. | [33] https://cytoscape.org/ |
| MOFA | R package (via Bioconductor) | All types (multi-omics) | Multi-Omics Factor Analysis enables the unsupervised integration of heterogeneous data sets via a generalization of principal components analysis. MOFA implements hidden factors of biological and technical sources of variability and represents integrated data in an interpretable low-dimensional form. | [34] https://github.com/bioFAM/MOFA |
| LUCID | R package (via CRAN) | Mainly genomics and metabolomics; integration of phenotypic data | Uses Latent Unknown Clusters with Integrated Data models to distinguish unique genomic, exposure and informative biomarkers or omics effects. Latent underlying relationships with phenotypic traits are estimated in cluster estimations using directed acyclic graphs (DAG). Prediction of phenotypes possible. Visualization of data integration with Sankey diagrams. | [35] https://github.com/USCbiostats/LUCIDus |
| MultiDataSet | R package (via Bioconductor) | Epigenomics, transcriptomics; assay data, feature data, phenotypic data stored in single object | Does not provide tools for analysis itself, but constructs an R data-storage object that contains multiple data sets, making managing and subsetting multiple and non-complete data sets possible. This data set can be plugged in to other R packages for analysis, for instance for multivariate co-inertia analysis (MCIA in omicade4) or clustering of multiple tables (in iClusterPlus) | [36] https://bioconductor.org/packages/release/bioc/html/MultiDataSet.html |
| Logicome Profiler | standalone Unix software | Applied to genomics and metagenomics, but applicable to any omics data | Detects statistically significant triplet logic relationships from a binary matrix dataset (indicating connection, for instance co-occurrence, co-expression). Applies Logic Analysis of Phylogenetic Profiles (LAPP) method, which is based on normalized mutual information, to phylogenetic profiling data, but also applicable to gene co-expression and pathway data. | [37] https://github.com/fukunagatsu/LogicomeProfiler |
| CoCoNet | R package (via Github) | Integration of GWAS and gene-expression data | COmposite likelihood-based COvariance regression NETwork model to identify trait-relevant tissues or cell types. Uses covariance regression network models to express gene-level effect measurements for a given GWAS trait as a function of the tissue-specific co-expression adjacency matrix. | [38] http://www.xzlab.org/software.html |
| NEO | R package (via CRAN) | Integration of GWAS and gene-expression data | Network Edge Orienting infers directed gene networks by integrating gene-expression data with genetic marker data and compares them with structural equation models | [39] https://horvath.genetics.ucla.edu/html/aten/NEO/ |
| WGCNA | R package (via CRAN) | Mainly gene-expression data, but can be applied to other omics | Weighted Gene Co-expression Network Analysis is used to find clusters, relating modules to one another and to external sample traits and calculates module membership measures. This approach facilitates gene screening and the identification of biomarkers. | [40,41] https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/ |
| DIABLO in mixOmics | R package (via Bioconductor) | All types (multi-omics) | Multivariate methods to analyse and visualize high-dimensional datasets (number of variables larger than number of samples). Complementary information from several data sets measured on the same N individuals, but across multiple omics data sets is combined to gain a better understanding of the interplay between the different levels of data that are measured (‘N-integration’). Data dimensions are reduced by applying sparse generalized canonical correlation analysis (SGCCA). | [42,43,44] http://mixomics.org/mixdiablo |
| Omics Type | Molecule Type | Molecule Name | Biofluid/Tissue | Description | Reference |
|---|---|---|---|---|---|
| epigenomics | DNA methylation | BCL-2 and RORA PPIEL | postmortem brains and peripheral blood cells | hypermethylation of BCL-2 and RORA genes in patients with autism; hypomethylation of PPIEL in bipolar disorder; hypermethylation of genes involved in brain development and tryptophan metabolism | [78] |
| DNA methylation | HTR1A, S-COMT, BDNF1 HTR1E, COMTD1 and MB-COMT | peripheral blood cells | peripheral epigenetic biomarkers of schizophrenia; hypermethylation of HTR1A, S-COMT, BDNF 1 hypomethylation of HTR1E, COMTD1 and MB-COMT | [79] | |
| DNA methylation | VWF and LRRC32 | hippocampus | reduced cognition in pigs in response to early life environmental insults (infection with porcine reproductive and respiratory syndrome virus) is associated with differential methylation and differential gene expression. VWF and LRRC32 are implicated in blood brain barrier permeability and regulatory T-cell activation, respectively. | [73] | |
| transcriptomics | miRNA | miR-24-2-5p, miR-27a-3p, miR-30e-5p, miR-3590-3p, miR-362-3p, and miR-532-5p | blood | pre-challenge circulating miRNAs reflect resilience or vulnerability to chronic social defeat in rats | [74] |
| miRNA | mir-132 | diverse tissues and fluids | associated with post-traumatic stress disorder in humans and animal models in a systematic review; lack of specificity | [75] | |
| miRNA | miR-19b, miR-27b, and miR-365 | saliva | concentrations greater in pigs that received no anti-inflammatory treatment after tail docking than in pigs that received treatment | [76] | |
| miRNA | range of circulating extra-cellular miRNAs | plasma | after feed deprivation in chicken lines selected for high and low residual feed intake, 23 and 19 miRNAs were found to be differentially expressed between feeding conditions and lines (indicating influence of genetic background), respectively. | [80] | |
| miRNA | range of circulating extra-cellular miRNAs | plasma | miRNA profiles were different between age classes (26 miRNAs) and lines (5 miRNAs) in dairy cattle. Three miRNAs negatively associated with telomere length, but positively with milk fat yield, mastitis and lameness. | [81] | |
| mRNA | profile | dorsal root ganglia | 3000 genes were differentially regulated between docked and undocked pigs | [56] | |
| mRNA | Pyruvate dehydrogenase (PDK4), heat shock (e.g. HSPB1) and oxidative (e.g. COX1) genes | longissimus dorsi muscle | up-regulated in the muscle of pigs under heat stress, reflecting the transition from glycolysis to fatty acid oxidation during chronic exposure to HS | [53] | |
| mRNA | profile | liver | A list of genes dose-dependently regulated by glucocorticoids as biomarkers of stress action | [82] | |
| proteomics | APP | Pig Major Acute Phase protein | serum | 7-fold increase in pigs after road transport | [58] |
| protein | GRP94 | liver | of critical importance at the onset of innate immune response, in pigs under HS. induces an inflammatory response, causing hepatocytes to synthesise haptoglobin (HP) and α-1-antichymotrypsin 2 precursor (SERPINA3) to maintain cell integrity. | [54] | |
| protein | lactate dehydrogenase (LDH) | saliva | significantly increased in the saliva of pigs restrained with a nose snare and in pigs with lameness. (LDH follows adrenaline production) | [57] | |
| protein | haptoglobin protegrin-3 and galectin-1 β-actin | blood serum | transition of sows from group to individual confined housing caused increase; indicates activation of immune defence and cell damage; indicates synthesis of stress-response hormones | [47] | |
| metabolomics | metabolite | 4,8-dimetil-nonanoyl carnitine | mesenteric adipose tissue | accumulation of 4,8-dimetil-nonanoyl carnitine, an intermediary of fatty acid oxidation, in this tissue of heat stressed pigs | [55] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasper, C.; Ribeiro, D.; Almeida, A.M.d.; Larzul, C.; Liaubet, L.; Murani, E. Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs. Genes 2020, 11, 920. https://doi.org/10.3390/genes11080920
Kasper C, Ribeiro D, Almeida AMd, Larzul C, Liaubet L, Murani E. Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs. Genes. 2020; 11(8):920. https://doi.org/10.3390/genes11080920
Chicago/Turabian StyleKasper, Claudia, David Ribeiro, André M. de Almeida, Catherine Larzul, Laurence Liaubet, and Eduard Murani. 2020. "Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs" Genes 11, no. 8: 920. https://doi.org/10.3390/genes11080920