Research Progress on Biological Accumulation, Detection and Inactivation Technologies of Norovirus in Oysters
Abstract
:1. Introduction
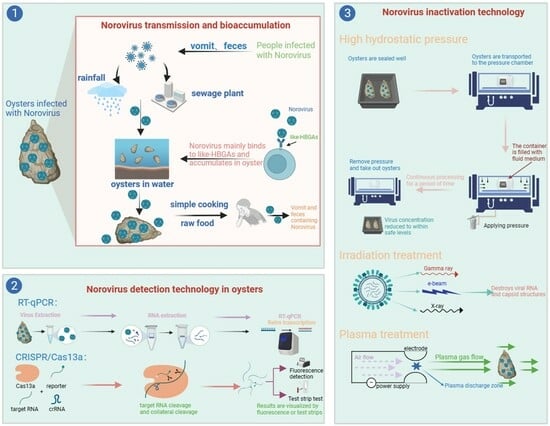
2. NoVs Contamination in Oysters
3. Bioaccumulation Mechanism of NoVs in Oysters
3.1. Oyster Feeding Mechanism
3.2. Bioaccumulation and Persistence of NoVs in Oysters
3.3. Inhibition of NoVs Binding to HBGA-Like Molecules
4. Potential Factors Affecting the Bioaccumulation of NoVs in Oysters
4.1. Temperature and Precipitation
4.2. Water Pollution
5. Detection Methods
6. Inactivation Methods
6.1. HHP
Influencing Factors
6.2. Irradiation Processing
6.3. Plasma Processing
7. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Atmar, R.L. Noroviruses: State of the Art. Food Environ. Virol. 2010, 2, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Ai, J.; Bao, C.; Zhang, J.; Wu, Q.; Zhu, L.; Hu, J.; Xing, Z. Evolution of the GII.3 P12 Norovirus from 2010 to 2019 in Jiangsu, China. Gut Pathog. 2021, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, Z. Detection and forecasting of oyster norovirus outbreaks: Recent advances and future perspectives. Mar. Environ. Res. 2012, 80, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.-W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Tong, L.; Wang, S.; Liu, N.; Zhao, F.; Sun, Y.; Sun, G.; Zhou, D. Gut Microbiota and Transcriptomics Reveal the Effect of Human Norovirus Bioaccumulation on Oysters (Crassostrea gigas). Microbiol. Spectr. 2022, 10, e0016122. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef]
- Lou, F.; Neetoo, H.; Chen, H.; Li, J. High Hydrostatic Pressure Processing: A Promising Nonthermal Technology to Inactivate Viruses in High-Risk Foods. Annu. Rev. Food Sci. Technol. 2015, 6, 389–409. [Google Scholar] [CrossRef]
- Shi, D.; Ma, H.; Miao, J.; Liu, W.; Yang, D.; Qiu, Z.; Shen, Z.; Yin, J.; Yang, Z.; Wang, H.; et al. Levels of human Rotaviruses and Noroviruses GII in urban rivers running through the city mirror their infection prevalence in populations. Sci. Total Environ. 2021, 754, 142203. [Google Scholar] [CrossRef]
- Tang, M.; Liao, N.; Tian, P.; Shen, K.; Liu, C.; Ruan, L.; Wu, G. Use of bentonite-coated activated carbon for improving the sensitivity of RT-qPCR detection of norovirus from vegetables and fruits: The ISO 15216-1:2017 standard method extension. Food Microbiol. 2023, 110, 104165. [Google Scholar] [CrossRef]
- Li, Y.; Xue, L.; Gao, J.; Cai, W.; Zhang, Z.; Meng, L.; Miao, S.; Hong, X.; Xu, M.; Wu, Q.; et al. A systematic review and meta-analysis indicates a substantial burden of human noroviruses in shellfish worldwide, with GII.4 and GII.2 being the predominant genotypes. Food Microbiol. 2023, 109, 104140. [Google Scholar] [CrossRef]
- Hassard, F.; Sharp, J.H.; Taft, H.; LeVay, L.; Harris, J.P.; McDonald, J.E.; Tuson, K.; Wilson, J.; Jones, D.L.; Malham, S.K. Critical Review on the Public Health Impact of Norovirus Contamination in Shellfish and the Environment: A UK Perspective. Food Environ. Virol. 2017, 9, 123–141. [Google Scholar] [CrossRef]
- Nagasawa, K.; Matsushima, Y.; Motoya, T.; Mizukoshi, F.; Ueki, Y.; Sakon, N.; Murakami, K.; Shimizu, T.; Okabe, N.; Nagata, N.; et al. Phylogeny and Immunoreactivity of Norovirus GII.P16-GII.2, Japan, Winter 2016–2017. Emerg. Infect. Dis. 2018, 24, 144–148. [Google Scholar] [CrossRef]
- El Moqri, N.; El Mellouli, F.; Hassou, N.; Benhafid, M.; Abouchoaib, N.; Etahiri, S. Norovirus Detection at Oualidia Lagoon, a Moroccan Shellfish Harvesting Area, by Reverse Transcription PCR Analysis. Food Environ. Virol. 2019, 11, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Miura, F.; Matsuyama, R.; Nishiura, H. Estimating the Asymptomatic Ratio of Norovirus Infection During Foodborne Outbreaks with Laboratory Testing in Japan. J. Epidemiol. 2018, 28, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Thebault, A.; Teunis, P.F.M.; Le Pendu, J.; Le Guyader, F.S.; Denis, J.-B. Infectivity of GI and GII noroviruses established from oyster related outbreaks. Epidemics 2013, 5, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Langlet, J.; Kaas, L.; Greening, G. Binding-Based RT-qPCR Assay to Assess Binding Patterns of Noroviruses to Shellfish. Food Environ. Virol. 2015, 7, 88–95. [Google Scholar] [CrossRef]
- Lees, D. Viruses and bivalve shellfish. Int. J. Food Microbiol. 2000, 59, 81–116. [Google Scholar] [CrossRef]
- Gyawali, P.; Fletcher, G.C.; McCoubrey, D.-J.; Hewitt, J. Norovirus in shellfish: An overview of post-harvest treatments and their challenges. Food Control 2019, 99, 171–179. [Google Scholar] [CrossRef]
- Morozov, V.; Hanisch, F.-G.; Wegner, K.M.; Schroten, H. Pandemic GII.4 Sydney and Epidemic GII.17 Kawasaki308 Noroviruses Display Distinct Specificities for Histo-Blood Group Antigens Leading to Different Transmission Vector Dynamics in Pacific Oysters. Front. Microbiol. 2018, 9, 2826. [Google Scholar] [CrossRef]
- Maalouf, H.; Zakhour, M.; Le Pendu, J.; Le Saux, J.-C.; Atmar, R.L.; Le Guyader, F.S. Distribution in Tissue and Seasonal Variation of Norovirus Genogroup I and II Ligands in Oysters. Appl. Environ. Microbiol. 2010, 76, 5621–5630. [Google Scholar] [CrossRef]
- Maalouf, H.; Schaeffer, J.; Parnaudeau, S.; Le Pendu, J.; Atmar, R.L.; Crawford, S.E.; Le Guyader, F.S. Strain-Dependent Norovirus Bioaccumulation in Oysters. Appl. Environ. Microbiol. 2011, 77, 3189–3196. [Google Scholar] [CrossRef] [PubMed]
- Lowther, J.A.; Henshilwood, K.; Lees, D.N. Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semiquantitative real-time reverse transcription PCR. J. Food Prot. 2008, 71, 1427–1433. [Google Scholar] [CrossRef]
- Tian, P.; Engelbrektson, A.L.; Mandrell, R.E. Seasonal tracking of histo-blood group antigen expression and norovirus binding in oyster gastrointestinal cells. J. Food Prot. 2008, 71, 1696–1700. [Google Scholar] [CrossRef] [PubMed]
- Guix, S.; Pinto, R.M.; Bosch, A. Final Consumer Options to Control and Prevent Foodborne Norovirus Infections. Viruses 2019, 11, 333. [Google Scholar] [CrossRef]
- Farkas, K.; Cooper, D.M.; McDonald, J.E.; Malham, S.K.; de Rougemont, A.; Jones, D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018, 634, 1174–1183. [Google Scholar] [CrossRef]
- Woods, J.W.; Calci, K.R.; Marchant-Tambone, J.G.; Burkhardt, W., III. Detection and molecular characterization of norovirus from oysters implicated in outbreaks in the US. Food Microbiol. 2016, 59, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Brake, F.; Ross, T.; Holds, G.; Kiermeier, A.; McLeod, C. A survey of Australian oysters for the presence of human noroviruses. Food Microbiol. 2014, 44, 264–270. [Google Scholar] [CrossRef]
- La Bella, G.; Martella, V.; Basanisi, M.G.; Nobili, G.; Terio, V.; La Salandra, G. Food-Borne Viruses in Shellfish: Investigation on Norovirus and HAV Presence in Apulia (SE Italy). Food Environ. Virol. 2017, 9, 179–186. [Google Scholar] [CrossRef]
- Kittigul, L.; Thamjaroen, A.; Chiawchan, S.; Chavalitshewinkoon-Petmitr, P.; Pombubpa, K.; Diraphat, P. Prevalence and Molecular Genotyping of Noroviruses in Market Oysters, Mussels, and Cockles in Bangkok, Thailand. Food Environ. Virol. 2016, 8, 133–140. [Google Scholar] [CrossRef]
- Lowther, J.A.; Gustar, N.E.; Powell, A.L.; O’Brien, S.; Lees, D.N. A One-Year Survey of Norovirus in UK Oysters Collected at the Point of Sale. Food Environ. Virol. 2018, 10, 278–287. [Google Scholar] [CrossRef]
- Sun, Z.; Niu, P.; Jin, M.; Gao, Q.; Wang, J.; Ma, X. Detection and Genetic Correlation Analysis of Diarrhea Cases and Norovirus in Oysters in Yantai, China. Front. Public Health 2022, 10, 819890. [Google Scholar] [CrossRef]
- Seo, D.J.; Lee, M.H.; Son, N.R.; Seo, S.; Lee, K.B.; Wang, X.; Choi, C. Seasonal and regional prevalence of norovirus, hepatitis A virus, hepatitis E virus, and rotavirus in shellfish harvested from South Korea. Food Control 2014, 41, 178–184. [Google Scholar] [CrossRef]
- Suffredini, E.; Le, Q.H.; Di Pasquale, S.; Pham, T.D.; Vicenza, T.; Losardo, M.; To, K.A.; De Medici, D. Occurrence and molecular characterization of enteric viruses in bivalve shellfish marketed in Vietnam. Food Control 2020, 108, 106828. [Google Scholar] [CrossRef]
- Loutreul, J.; Cazeaux, C.; Levert, D.; Nicolas, A.; Vautier, S.; Le Sauvage, A.L.; Perelle, S.; Morin, T. Prevalence of Human Noroviruses in Frozen Marketed Shellfish, Red Fruits and Fresh Vegetables. Food Environ. Virol. 2014, 6, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.T.H.; Ho, S.X.; Chu, J.J.H.; Li, D. Application of virome capture sequencing in shellfish sold at retail level in Singapore. Lett. Appl. Microbiol. 2021, 73, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhao, F.; Tong, L.; Wang, S.; Zhou, D. Contamination, bioaccumulation mechanism, detection, and control of human norovirus in bivalve shellfish: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 8972–8985. [Google Scholar] [CrossRef]
- Provost, K.; Dancho, B.A.; Ozbay, G.; Anderson, R.S.; Richards, G.P.; Kingsley, D.H. Hemocytes Are Sites of Enteric Virus Persistence within Oysters. Appl. Environ. Microbiol. 2011, 77, 8360–8369. [Google Scholar] [CrossRef]
- Tame, A.; Ozawa, G.; Maruyama, T.; Yoshida, T. Morphological and functional characterization of hemocytes from two deep-sea vesicomyid clams Phreagena okutanii and Abyssogena phaseoliformis. Fish Shellfish Immunol. 2018, 74, 281–294. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D. Specific Interactions between Human Norovirus and Environmental Matrices: Effects on the Virus Ecology. Viruses 2019, 11, 224. [Google Scholar] [CrossRef]
- McLeod, C.; Hay, B.; Grant, C.; Greening, G.; Day, D. Inactivation and elimination of human enteric viruses by Pacific oysters. J. Appl. Microbiol. 2009, 107, 1809–1818. [Google Scholar] [CrossRef]
- Le Mennec, C.; Parnaudeau, S.; Rumebe, M.; Le Saux, J.-C.; Piquet, J.-C.; Le Guyader, S.F. Follow-Up of Norovirus Contamination in an Oyster Production Area Linked to Repeated Outbreaks. Food Environ. Virol. 2017, 9, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.P.; McLeod, C.; Le Guyader, F.S. Processing Strategies to Inactivate Enteric Viruses in Shellfish. Food Environ. Virol. 2010, 2, 183–193. [Google Scholar] [CrossRef]
- Le Guyader, F.S.; Le Saux, J.-C.; Ambert-Balay, K.; Krol, J.; Serais, O.; Parnaudeau, S.; Giraudon, H.; Delmas, G.; Pommepuy, M.; Pothier, P.; et al. Aichi Virus, Norovirus, Astrovirus, Enterovirus, and Rotavirus Involved in Clinical Cases from a French Oyster-Related Gastroenteritis Outbreak. J. Clin. Microbiol. 2008, 46, 4011–4017. [Google Scholar] [CrossRef] [PubMed]
- Nasir, W.; Frank, M.; Kunze, A.; Bally, M.; Parra, F.; Nyholm, P.-G.; Hook, F.; Larson, G. Histo-Blood Group Antigen Presentation Is Critical for Binding of Norovirus VLP to Glycosphingolipids in Model Membranes. ACS Chem. Biol. 2017, 12, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Le Guyader, F.S.; Loisy, F.; Atmar, R.L.; Hutson, A.M.; Estes, M.K.; Ruvoen-Clouet, N.; Pommepuy, M.; Le Pendu, J. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 2006, 12, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wu, Q.; Kou, X.; Yao, L.; Zhang, J. Distribution of norovirus in oyster tissues. J. Appl. Microbiol. 2008, 105, 1966–1972. [Google Scholar] [CrossRef]
- Su, L.; Ma, L.; Liu, H.; Zhao, F.; Su, Z.; Zhou, D. Presence and Distribution of Histo-Blood Group Antigens in Pacific Oysters and the Effects of Exposure to Noroviruses GI.3 and GII.4 on Their Expression. J. Food Prot. 2018, 81, 1783–1790. [Google Scholar] [CrossRef]
- Le Guyader, F.S.; Atmar, R.L.; Le Pendu, J. Transmission of viruses through shellfish: When specific ligands come into play. Curr. Opin. Virol. 2012, 2, 103–110. [Google Scholar] [CrossRef]
- Schroten, H.; Hanisch, F.-G.; Hansman, G.S. Human Norovirus Interactions with Histo-Blood Group Antigens and Human Milk Oligosaccharides. J. Virol. 2016, 90, 5855–5859. [Google Scholar] [CrossRef]
- Patin, N.V.; Pena-Gonzalez, A.; Hatt, J.K.; Moe, C.; Kirby, A.; Konstantinidis, K.T. The Role of the Gut Microbiome in Resisting Norovirus Infection as Revealed by a Human Challenge Study. Mbio 2020, 11, e02634-20. [Google Scholar] [CrossRef]
- Li, Y.; Gao, J.; Xue, L.; Shang, Y.; Cai, W.; Xie, X.; Jiang, T.; Chen, H.; Zhang, J.; Wang, J.; et al. Determination of Antiviral Mechanism of Centenarian Gut-Derived Limosilactobacillus fermentum Against Norovirus. Front. Nutr. 2022, 9, 812623. [Google Scholar] [CrossRef] [PubMed]
- Gorji, M.E.; Tan, M.T.H.; Li, D. Influence of fucosidase-producing bifidobacteria on the HBGA antigenicity of oyster digestive tissue and the associated norovirus binding. Int. J. Food Microbiol. 2021, 340, 109058. [Google Scholar] [CrossRef]
- Chenar, S.S.; Deng, Z. Environmental indicators for human norovirus outbreaks. Int. J. Environ. Health Res. 2017, 27, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.J.A.; Lees, D.N. Environmental Transmission of Human Noroviruses in Shellfish Waters. Appl. Environ. Microbiol. 2014, 80, 3552–3561. [Google Scholar] [CrossRef]
- Parikh, M.P.; Vandekar, S.; Moore, C.; Thomas, L.; Britt, N.; Piya, B.; Stewart, L.S.; Batarseh, E.; Hamdan, L.; Cavallo, S.J.; et al. Temporal and Genotypic Associations of Sporadic Norovirus Gastroenteritis and Reported Norovirus Outbreaks in Middle Tennessee, 2012–2016. Clin. Infect. Dis. 2020, 71, 2398–2404. [Google Scholar] [CrossRef]
- Rince, A.; Baliere, C.; Hervio-Heath, D.; Cozien, J.; Lozach, S.; Parnaudeau, S.; Le Guyader, F.S.; Le Hello, S.; Giard, J.-C.; Sauvageot, N.; et al. Occurrence of Bacterial Pathogens and Human Noroviruses in Shellfish-Harvesting Areas and Their Catchments in France. Front. Microbiol. 2018, 9, 2443. [Google Scholar] [CrossRef]
- Choi, C.; Kingsley, D.H. Temperature-Dependent Persistence of Human Norovirus within Oysters (Crassostrea virginica). Food Environ. Virol. 2016, 8, 141–147. [Google Scholar] [CrossRef]
- Polo, D.; Varela, M.F.; Romalde, J.L. Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas. Int. J. Food Microbiol. 2015, 193, 43–50. [Google Scholar] [CrossRef]
- Alvarez, I.; Lorenzo, M.N.; deCastro, M. Analysis of chlorophyll a concentration along the Galician coast: Seasonal variability and trends. ICES J. Mar. Sci. 2012, 69, 728–738. [Google Scholar] [CrossRef]
- Gentry, J.; Vinje, J.; Guadagnoli, D.; Lipp, E.K. Norovirus Distribution within an Estuarine Environment. Appl. Environ. Microbiol. 2009, 75, 5474–5480. [Google Scholar] [CrossRef]
- Eregno, F.E.; Tryland, I.; Tjomsland, T.; Myrmel, M.; Robertson, L.; Heistad, A. Quantitative microbial risk assessment combined with hydrodynamic modelling to estimate the public health risk associated with bathing after rainfall events. Sci. Total Environ. 2016, 548, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.; Fitzhenry, K.; O’Flaherty, V.; Dore, W.; Keaveney, S.; Cormican, M.; Rowan, N.; Clifford, E. Detection, fate and inactivation of pathogenic norovirus employing settlement and UV treatment in wastewater treatment facilities. Sci. Total Environ. 2016, 568, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, M.A.; Bradbury, K.R.; Alexander, E.C., Jr.; Kolberg, R.J.; Alexander, S.C.; Archer, J.R.; Braatz, L.A.; Forest, B.M.; Green, J.A.; Spencer, S.K. Norovirus Outbreak Caused by a New Septic System in a Dolomite Aquifer. Ground Water 2011, 49, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.A.; Gundy, P.M.; Rijal, G.K.; Gerba, C.P. The Impact of Combined Sewage Overflows on the Viral Contamination of Receiving Waters. Food Environ. Virol. 2012, 4, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, J.; Treguier, C.; Piquet, J.-C.; Gachelin, S.; Cochennec-Laureau, N.; Le Saux, J.-C.; Garry, P.; Le Guyader, F.S. Improving the efficacy of sewage treatment decreases norovirus contamination in oysters. Int. J. Food Microbiol. 2018, 286, 1–5. [Google Scholar] [CrossRef]
- Campos, C.J.A.; Avant, J.; Lowther, J.; Till, D.; Lees, D.N. Human norovirus in untreated sewage and effluents from primary, secondary and tertiary treatment processes. Water Res. 2016, 103, 224–232. [Google Scholar] [CrossRef]
- Ibrahim, C.; Hammami, S.; Khelifi, N.; Pothier, P.; Hassen, A. The Effectiveness of Activated Sludge Procedure and UV-C(254)in Norovirus Inactivation in a Tunisian Industrial Wastewater Treatment Plant. Food Environ. Virol. 2020, 12, 250–259. [Google Scholar] [CrossRef]
- Prado, T.; Bruni, A.d.C.; Funada Barbosa, M.R.; Garcia, S.C.; Moreno, L.Z.; Zanoli Sato, M.I. Noroviruses in raw sewage, secondary effluents and reclaimed water produced by sand-anthracite filters and membrane bioreactor/reverse osmosis system. Sci. Total Environ. 2019, 646, 427–437. [Google Scholar] [CrossRef]
- Sarmento, S.K.; Guerra, C.R.; Malta, F.C.; Coutinho, R.; Miagostovich, M.P.; Fumian, T.M. Human norovirus detection in bivalve shellfish in Brazil and evaluation of viral infectivity using PMA treatment. Mar. Pollut. Bull. 2020, 157, 111315. [Google Scholar] [CrossRef]
- Wang, N.; Pan, G.; Liu, P.; Rong, S.; Gao, Z.; Li, Q. Advances and Future Perspective on Detection Technology of Human Norovirus. Pathogens 2021, 10, 1383. [Google Scholar] [CrossRef]
- Gyawali, P.; Sanjaya, K.C.; Beale, D.J.; Hewitt, J. Current and Emerging Technologies for the Detection of Norovirus from Shellfish. Foods 2019, 8, 187. [Google Scholar] [CrossRef]
- Stals, A.; Uyttendaele, M.; Van Coillie, E. The Need for Harmonization in Detection of Human Noroviruses in Food. J. AOAC Int. 2013, 96, 998–1005. [Google Scholar] [CrossRef]
- Manuel, C.S.; Moore, M.D.; Jaykus, L.-A. Predicting human norovirus infectivity—Recent advances and continued challenges. Food Microbiol. 2018, 76, 337–345. [Google Scholar] [CrossRef]
- Persson, S.; Eriksson, R.; Lowther, J.; Ellstrom, P.; Simonsson, M. Comparison between RT droplet digital PCR and RT real-time PCR for quantification of noroviruses in oysters. Int. J. Food Microbiol. 2018, 284, 73–83. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2020, 15, 1311. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Yang, X.; Zhan, W.; Tang, Y.; Wei, M.; Chen, K.; Liu, P.; Xie, J.; Zhang, C.; Zhao, H.; et al. Development of a rapid and accurate CRISPR/Cas13-based diagnostic test for GII.4 norovirus infection. Front. Microbiol. 2022, 13, 912315. [Google Scholar] [CrossRef] [PubMed]
- Araud, E.; DiCaprio, E.; Ma, Y.; Lou, F.; Gao, Y.; Kingsley, D.; Hughes, J.H.; Li, J. Thermal Inactivation of Enteric Viruses and Bioaccumulation of Enteric Foodborne Viruses in Live Oysters (Crassostrea virginica). Appl. Environ. Microbiol. 2016, 82, 2086–2099. [Google Scholar] [CrossRef] [PubMed]
- Croci, L.; Suffredini, E.; Di Pasquale, S.; Cozzi, L. Detection of Norovirus and Feline Calicivirus in spiked molluscs subjected to heat treatments. Food Control 2012, 25, 17–22. [Google Scholar] [CrossRef]
- Tong, L.; Ding, G.; Yang, M.; Su, L.; Wang, S.; Wang, Y.; Zheng, L.; Zhou, D.; Zhao, F. High-hydrostatic-pressure inactivation of GI.5 and GII.4 human norovirus and effects on the physical, chemical, and taste characteristics of oyster (Crassostrea virginica). LWT 2023, 176, 114554. [Google Scholar] [CrossRef]
- Ahmed, H.; Maunula, L.; Korhonen, J. Reduction of Norovirus in Foods by Nonthermal Treatments: A Review. J. Food Prot. 2020, 83, 2053–2073. [Google Scholar] [CrossRef] [PubMed]
- Rowan, N.J. Current decontamination challenges and potentially complementary solutions to safeguard the vulnerable seafood industry from recalcitrant human norovirus in live shellfish: Quo Vadis? Sci. Total Environ. 2023, 874, 162380. [Google Scholar] [CrossRef] [PubMed]
- Usman, I.; Afzaal, M.; Imran, A.; Saeed, F.; Afzal, A.; Ashfaq, I.; Shah, Y.A.; Islam, F.; Azam, I.; Tariq, I.; et al. Recent updates and perspectives of plasma in food processing: A review. Int. J. Food Prop. 2023, 26, 552–566. [Google Scholar] [CrossRef]
- Praveen, C.; Dancho, B.A.; Kingsley, D.H.; Calci, K.R.; Meade, G.K.; Mena, K.D.; Pillai, S.D. Susceptibility of Murine Norovirus and Hepatitis A Virus to Electron Beam Irradiation in Oysters and Quantifying the Reduction in Potential Infection Risks. Appl. Environ. Microbiol. 2013, 79, 3796–3801. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Park, S.Y.; Rui, M.-L.; Ha, S.-D. Effects of electron beam irradiation on murine norovirus-1 in abalone (Haliotis discus hannai) meat and viscera. LWT Food Sci. Technol. 2017, 86, 611–618. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, S.; Nannapaneni, R.; Coker, R.; Haque, Z.; Mahmoud, B.S.M. The efficacy of X-ray doses on murine norovirus-1 (MNV-1) in pure culture, half-shell oyster, salmon sushi, and tuna salad. Food Control 2016, 64, 77–80. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, S.; Nannapaneni, R.; Zhang, Y.; Coker, R.; Mahmoud, B.S.M. The effects of X-ray treatments on bioaccumulated murine norovirus1 (MNV-1) and survivability, inherent microbiota, color, and firmness of Atlantic oysters (Crassostrea virginica) during storage at 5 degrees C for 20 days. Food Control 2017, 73, 1189–1194. [Google Scholar] [CrossRef]
- Takahashi, M.; Okakura, Y.; Takahashi, H.; Yamane, H.; Akashige, S.; Kuda, T.; Kimura, B. Evaluation of Inactivation of Murine Norovirus in Inoculated Shell Oysters by High Hydrostatic Pressure Treatment. J. Food Prot. 2019, 82, 2169–2173. [Google Scholar] [CrossRef]
- Li, X.; Ye, M.; Neetoo, H.; Golovan, S.; Chen, H. Pressure inactivation of Tulane virus, a candidate surrogate for human norovirus and its potential application in food industry. Int. J. Food Microbiol. 2013, 162, 37–42. [Google Scholar] [CrossRef]
- Ye, M.; Li, X.; Kingsley, D.H.; Jiang, X.; Chen, H. Inactivation of Human Norovirus in Contaminated Oysters and Clams by High Hydrostatic Pressure. Appl. Environ. Microbiol. 2014, 80, 2248–2253. [Google Scholar] [CrossRef]
- Ye, M.; Lingham, T.; Huang, Y.; Ozbay, G.; Ji, L.; Karwe, M.; Chen, H. Effects of High-Hydrostatic Pressure on Inactivation of Human Norovirus and Physical and Sensory Characteristics of Oysters. J. Food Sci. 2015, 80, M1330–M1335. [Google Scholar] [CrossRef]
- Li, X.; Chen, H. Evaluation of the Porcine Gastric Mucin Binding Assay for High-Pressure-Inactivation Studies Using Murine Norovirus and Tulane Virus. Appl. Environ. Microbiol. 2015, 81, 515–521. [Google Scholar] [CrossRef]
- Bozkurt, H.; Kim-Yen, P.-T.; van Ogtrop, F.; Bell, T.; McConchie, R. Outbreaks, occurrence, and control of norovirus and hepatitis a virus contamination in berries: A review. Crit. Rev. Food Sci. Nutr. 2021, 61, 116–138. [Google Scholar] [CrossRef]
- Pereira, R.N.; Vicente, A.A. Environmental impact of novel thermal and non-thermal technologies in food processing. Food Res. Int. 2010, 43, 1936–1943. [Google Scholar] [CrossRef]
- Chawla, R.; Patil, G.R.; Singh, A.K. High hydrostatic pressure technology in dairy processing: A review. J. Food Sci. Technol. 2011, 48, 260–268. [Google Scholar] [CrossRef]
- Leon, J.S.; Kingsley, D.H.; Montes, J.S.; Richards, G.P.; Lyon, G.M.; Abdulhafid, G.M.; Seitz, S.R.; Fernandez, M.L.; Teunis, P.F.; Flick, G.J.; et al. Randomized, Double-Blinded Clinical Trial for Human Norovirus Inactivation in Oysters by High Hydrostatic Pressure Processing. Appl. Environ. Microbiol. 2011, 77, 5476–5482. [Google Scholar] [CrossRef]
- Kingsley, D.H. High Pressure Processing of Bivalve Shellfish and HPP’s Use as a Virus Intervention. Foods 2014, 3, 336–350. [Google Scholar] [CrossRef] [PubMed]
- Terio, V.; Tantillo, G.; Martella, V.; Di Pinto, P.; Buonavoglia, C.; Kingsley, D.H. High Pressure Inactivation of HAV within Mussels. Food Environ. Virol. 2010, 2, 83–88. [Google Scholar] [CrossRef]
- Truong, B.Q.; Buckow, R.; Stathopoulos, C.E.; Nguyen, M.H. Advances in High-Pressure Processing of Fish Muscles. Food Eng. Rev. 2015, 7, 109–129. [Google Scholar] [CrossRef]
- Lou, F.; Neetoo, H.; Chen, H.; Li, J. Inactivation of a Human Norovirus Surrogate by High-Pressure Processing: Effectiveness, Mechanism, and Potential Application in the Fresh Produce Industry. Appl. Environ. Microbiol. 2011, 77, 1862–1871. [Google Scholar] [CrossRef] [PubMed]
- DiCaprio, E.; Ye, M.; Chen, H.; Li, J. Inactivation of Human Norovirus and Tulane Virus by High Pressure Processing in Simple Mediums and Strawberry Puree. Front. Sustain. Food Syst. 2019, 3, 26. [Google Scholar] [CrossRef]
- Hewitt, J.; Rivera-Aban, M.; Greening, G.E. Evaluation of murine norovirus as a surrogate for human norovirus and hepatitis A virus in heat inactivation studies. J. Appl. Microbiol. 2009, 107, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Q.; Hoover, D.G.; Kingsley, D.H. Temperature and treatment time influence high hydrostatic pressure inactivation of feline calicivirus, a norovirus surrogate. J. Food Prot. 2005, 68, 2389–2394. [Google Scholar] [CrossRef]
- Kingsley, D.H.; Hollinian, D.R.; Calci, K.R.; Chen, H.; Flick, G.J. Inactivation of a norovirus by high-pressure processing. Appl. Environ. Microbiol. 2007, 73, 581–585. [Google Scholar] [CrossRef]
- Sido, R.F.; Huang, R.; Liu, C.; Chen, H. High hydrostatic pressure inactivation of murine norovirus and human noroviruses on green onions and in salsa. Int. J. Food Microbiol. 2017, 242, 1–6. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Kingsley, D.H. The influence of temperature, pH, and water immersion on the high hydrostatic pressure inactivation of GI.1 and GII.4 human noroviruses. Int. J. Food Microbiol. 2013, 167, 138–143. [Google Scholar] [CrossRef]
- Kunugi, S.; Tanaka, N. Cold denaturation of proteins under high pressure. Biochim. Biophys. Acta 2002, 1595, 329–344. [Google Scholar] [CrossRef]
- Balny, C.; Masson, P.; Heremans, K. High pressure effects on biological macromolecules: From structural changes to alteration of cellular processes. Biochim. Biophys. Acta 2002, 1595, 3–10. [Google Scholar] [CrossRef]
- Kingsley, D.H.; Chen, H. Aqueous matrix compositions and pH influence feline calicivirus inactivation by high pressure processing. J. Food Prot. 2008, 71, 1598–1603. [Google Scholar] [CrossRef]
- Lou, F.; Neetoo, H.; Li, J.; Chen, H.; Li, J. Lack of Correlation between Virus Barosensitivity and the Presence of a Viral Envelope during Inactivation of Human Rotavirus, Vesicular Stomatitis Virus, and Avian Metapneumovirus by High-Pressure Processing. Appl. Environ. Microbiol. 2011, 77, 8538–8547. [Google Scholar] [CrossRef]
- Baert, L.; Debevere, J.; Uyttendaele, M. The efficacy of preservation methods to inactivate foodborne viruses. Int. J. Food Microbiol. 2009, 131, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, D.H.; Chen, H. Influence of pH, salt, and temperature on pressure inactivation of hepatitis A virus. Int. J. Food Microbiol. 2009, 130, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Li, D.; Xu, J.; Wang, J.; Zhao, Y.; Li, Z.; Xue, C. Mechanism of inactivation of murine norovirus-1 by high pressure processing. Int. J. Food Microbiol. 2010, 137, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; DiCaprio, E.; Li, X.; Dai, X.; Ma, Y.; Hughes, J.; Chen, H.; Kingsley, D.H.; Li, J. Variable High-Pressure-Processing Sensitivities for Genogroup II Human Noroviruses. Appl. Environ. Microbiol. 2016, 82, 6037–6045. [Google Scholar] [CrossRef] [PubMed]
- Pi, X.; Yang, Y.; Sun, Y.; Wang, X.; Wan, Y.; Fu, G.; Li, X.; Cheng, J. Food irradiation: A promising technology to produce hypoallergenic food with high quality. Crit. Rev. Food Sci. Nutr. 2022, 62, 6698–6713. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.; Pavase, T.; Li, Z.; Liu, Y.; Wang, N. Evaluation of electron beam irradiation to reduce the IgE binding capacity of frozen shrimp tropomyosin. Food Agric. Immunol. 2017, 28, 189–201. [Google Scholar] [CrossRef]
- Feng, K.; Divers, E.; Ma, Y.; Li, J. Inactivation of a Human Norovirus Surrogate, Human Norovirus Virus-Like Particles, and Vesicular Stomatitis Virus by Gamma Irradiation. Appl. Environ. Microbiol. 2011, 77, 3507–3517. [Google Scholar] [CrossRef]
- Park, S.Y.; Ha, S.-D. Application of gamma radiation for the reduction of norovirus and the quality stability in optimally ripened cabbage kimchi. Food Res. Int. 2017, 100, 277–281. [Google Scholar] [CrossRef]
- DiCaprio, E.; Phantkankum, N.; Culbertson, D.; Ma, Y.; Hughes, J.H.; Kingsley, D.; Uribe, R.M.; Li, J. Inactivation of human norovirus and Tulane virus in simple media and fresh whole strawberries by ionizing radiation. Int. J. Food Microbiol. 2016, 232, 43–51. [Google Scholar] [CrossRef]
- Predmore, A.; Sanglay, G.C.; DiCaprio, E.; Li, J.; Uribe, R.M.; Lee, K. Electron beam inactivation of Tulane virus on fresh produce, and mechanism of inactivation of human norovirus surrogates by electron beam irradiation. Int. J. Food Microbiol. 2015, 198, 28–36. [Google Scholar] [CrossRef]
- Pankaj, S.K.; Wan, Z.; Keener, K.M. Effects of Cold Plasma on Food Quality: A Review. Foods 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Aboubakr, H.A.; Mor, S.K.; Higgins, L.; Armien, A.; Youssef, M.M.; Bruggeman, P.J.; Goyal, S.M. Cold argon-oxygen plasma species oxidize and disintegrate capsid protein of feline calicivirus. PLoS ONE 2018, 13, e0194618. [Google Scholar] [CrossRef] [PubMed]
- Filipic, A.; Gutierrez-Aguirre, I.; Primc, G.; Mozetic, M.; Dobnik, D. Cold Plasma, a New Hope in the Field of Virus Inactivation. Trends Biotechnol. 2020, 38, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Velebit, B.; Milojevic, L.; Baltic, T.; Grkovic, N.; Gummalla, S.; Velebit, M.; Skoko, I.; Mojsova, S.; Putnik, P. Efficacy of cold atmospheric plasma for inactivation of viruses on raspberries. Innov. Food Sci. Emerg. Technol. 2022, 81, 103121. [Google Scholar] [CrossRef]
- Aboubakr, H.A.; Parra, F.S.; Collins, J.; Bruggeman, P.; Goyal, S.M. In situ inactivation of human norovirus GII.4 by cold plasma: Ethidium monoazide (EMA)-coupled RT-qPCR underestimates virus reduction and fecal material suppresses inactivation. Food Microbiol. 2020, 85, 103307. [Google Scholar] [CrossRef]
- Choi, M.-S.; Jeon, E.B.; Kim, J.Y.; Choi, E.H.; Lim, J.S.; Choi, J.; Ha, K.S.; Kwon, J.Y.; Jeong, S.H.; Park, S.Y. Virucidal Effects of Dielectric Barrier Discharge Plasma on Human Norovirus Infectivity in Fresh Oysters (Crassostrea gigas). Foods 2020, 9, 1731. [Google Scholar] [CrossRef]
- Dong, S.; Niu, Y.; Wei, H.; Lin, Y.; Lu, X.; Yamashita, T.; Yu, K.; Takaki, K.; Yuan, C. Effect of super-chilling storage on maintenance of quality and freshness of the Pacific oyster (Crassostrea gigas). Food Qual. Saf. 2023, 7, fyad008. [Google Scholar] [CrossRef]


| Area | Period | Species | GI (%) | GII (%) | References |
|---|---|---|---|---|---|
| Australia | 2010–2011 | Oysters | - | 1.7 | [27] |
| Italy | January 2013–July 2015 | Mussels | 3.0 | 26.0 | [28] |
| Oysters | - | 1.0 | |||
| Thailand | August 2011–July 2012 | Mussels | 9.0 | 4.0 | [29] |
| Oysters | 10.0 | 2.0 | |||
| Clams | 4.0 | 2.0 | |||
| UK | March 2015–March 2016 | Oysters | 15.7 | 14.0 | [30] |
| China | March 2019 | Oysters | 6.6 | 21.2 | [31] |
| Republic of Korea | Janurary 2011–September 2011 | Oysters | 7.8 | 15.7 | [32] |
| Vietnam | October 2015–June 2016 | Oysters | 52.4 | 74.6 | [33] |
| Moroccan Atlantic coast | November 2015–February 2017 | Oysters | - | 7.0 | [13] |
| France | Janurary 2008–September 2008 | Mussels | 8.4 | 14.4 | [34] |
| Singapore | 2019–2020 | Oysters | 35.3 | 34.6 | [35] |
| NTP Technologies | NoV Types | Processing Conditions | Initial Concentration | Reduction (%) | References |
|---|---|---|---|---|---|
| e-beam irradiation | MNV-1 | 5 kGy | 4.9 log PFU/mL | 12 | [84] |
| e-beam irradiation | MNV-1 | 7 kGy | 6.06 log10 PFU/mL | 30 | [85] |
| X-Ray irradiation | MNV-1 | 4 kGy | 6.3 log PFU/mL | 59 | [86] |
| X-Ray irradiation | MNV-1 | 1 kGy | 4.3 log PFU/g | 16 | [87] |
| 2 kGy | 25 | ||||
| 3 kGy | 35 | ||||
| 4 kGy | 42 | ||||
| HHP | MNV-1 | 275 MPa, 2 min, 0 °C | 4.8 log PFU/oyster | 2 | [88] |
| 275 MPa, 5 min, 0 °C | 42 | ||||
| HHP | MNV-1 | 350 MPa, 2 min, 4 °C | 5.5 log10 PFU/g | 36 | [89] |
| HHP | HuNoV GII.4 | 300 MPa, 5 min, 6 °C | 4 log10 units | 72 | [90] |
| 400 MPa, 5 min, 6 °C | 90 | ||||
| HuNoV GI.1 | 300 MPa, 5 min, 6 °C | 17 | |||
| 400 MPa, 5 min, 6 °C | 32 | ||||
| HHP | HuNoV GII.4 | 350 MPa, 2 min, 0 °C | 4–5 log10 units | >84 | [91] |
| 350 MPa, 2 min, 25 °C | 72–90 | ||||
| HuNoV GI.1 | 500 MPa, 2 min, 0 °C | >86 | |||
| 500 MPa, 2 min, 25 °C | 16–20 | ||||
| HHP | HuNoV GII.4 | 350 MPa, 2 min, 21 °C, pH 4 | 6 log10 units | 30 | [92] |
| HuNoV GI.1 | 550 MPa, 2 min, 21 °C, pH 4 | 18 | |||
| HHP | HuNoV GII.4 | 200 MPa, 5 min, 5 °C | 8.95 × 105 log10 genome copies | 89 | [80] |
| HuNoV GI.5 | 4.39 × 105 log10 genome copies | 85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liang, M.; Zhao, F.; Su, L. Research Progress on Biological Accumulation, Detection and Inactivation Technologies of Norovirus in Oysters. Foods 2023, 12, 3891. https://doi.org/10.3390/foods12213891
Sun Y, Liang M, Zhao F, Su L. Research Progress on Biological Accumulation, Detection and Inactivation Technologies of Norovirus in Oysters. Foods. 2023; 12(21):3891. https://doi.org/10.3390/foods12213891
Chicago/Turabian StyleSun, Yiqiang, Meina Liang, Feng Zhao, and Laijin Su. 2023. "Research Progress on Biological Accumulation, Detection and Inactivation Technologies of Norovirus in Oysters" Foods 12, no. 21: 3891. https://doi.org/10.3390/foods12213891