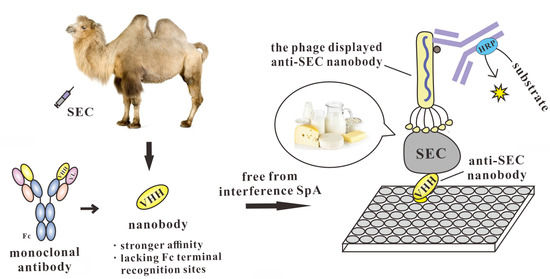
Development of a Double Nanobody-Based Sandwich Immunoassay for the Detecting Staphylococcal Enterotoxin C in Dairy Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bactrian Camel Immunization
2.3. Constructing the Phage Display Nb Library
2.4. Biopanning of the Phage Display Anti-SEC Nbs
2.5. Preparation of the Phage Display Nbs
2.6. The Purification and Expression of the Anti-SEC Nbs
2.7. Pairwise Selection for Sandwich Nbs-ELISA
2.8. Developing a Sandwich Nbs-ELISA for SEC Detection
2.9. Cross-Reactivity Examination
2.10. Analysis of the Spiked Dairy Product
3. Results
3.1. Anti-SEC Nb Library Construction
3.2. Biopanning of the Anti-SEC Nbs
3.3. Characterization of the Binding Properties of Anti-SEC Nbs
3.4. Pairwise Interaction Analysis
3.5. Sandwich Nbs-ELISA for SEC Detection
3.6. Cross-Reactivity Assay
3.7. Spiked Sample Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Lefebvre, D.; Blanco-Valle, K.; Feraudet-Tarisse, C.; Merda, D.; Simon, S.; Fenaille, F.; Hennekinne, J.-A.; Nia, Y.; Becher, F. Quantitative Determination of Staphylococcus aureus Enterotoxins Types A to I and Variants in Dairy Food Products by Multiplex Immuno-LC-MS/MS. J. Agric. Food Chem. 2021, 69, 2603–2610. [Google Scholar] [CrossRef]
- Etter, D.; Schelin, J.; Schuppler, M.; Johler, S. Staphylococcal enterotoxin C-an update on SEC variants, their structure and properties, and their role in foodborne intoxications. Toxins 2000, 12, 584. [Google Scholar] [CrossRef]
- Valihrach, L.; Alibayov, B.; Zdenkova, K.; Demnerova, K. Expression and production of staphylococcal enterotoxin C is substantially reduced in milk. Food Microbiol. 2014, 44, 54–59. [Google Scholar] [CrossRef]
- Benkerroum, N. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: An overview. Crit. Rev. Food Sci. Nutr. 2018, 58, 1943–1970. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Ali, H.A.; Gorish, B.M.T.; Ali, S.O.; Abdalrhim, E.S.A.; Mergani, M.H.; Elgadir, A.A.A.; Mohammed, S.K.; Ahmed, S.O.; Musa, N.A.; et al. Molecular Detection of Staphylococcal Enterotoxins and mecA Genes Products in Selected Food Samples Collected from Different Areas in Khartoum State. Int. J. Microbiol. 2021, 2021, 5520573. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, R.; Wang, S.; Yang, X.; Bai, Z.; Li, X.; Rong, Z.; Shen, B.; Wang, S. Magnetic quantum dot based lateral flow assay biosensor for multiplex and sensitive detection of protein toxins in food samples. Biosens. Bioelectron. 2019, 146, 111754. [Google Scholar] [CrossRef]
- Tonacini, J.; Stephan, D.; Vogel, G.; Avondet, M.A.; Kalman, F.; Crovadore, J.; Schnyder, B. Intact Staphylococcus enterotoxin SEB from culture supernatant detected by MALDI-TOF mass spectrometry. Toxins 2019, 11, 101. [Google Scholar] [CrossRef] [Green Version]
- Jayan, H.; Pu, H.; Sun, D.W. Recent development in rapid detection techniques for microorganism activities in food matrices using bio-recognition: A review. Trends Food Sci. Tech. 2020, 95, 233–246. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Wang, Z.; Jiang, H.; Lv, Z.; Shen, J.; Xia, G.; Wen, K. Universal simultaneous multiplex ELISA of small molecules in milk based on dual luciferases. Anal. Chim. Acta 2018, 1001, 125–133. [Google Scholar] [CrossRef]
- Abril, A.G.; Gonzalez-Villa, T.; Barros-Velázquez, J.; Cañas, B.; Sánchez-Pérez, A.; Calo-Mata, P.; Carrera, M. Staphylococcus aureus exotoxins and their detection in the dairy industry and mastitis. Toxins 2020, 12, 537. [Google Scholar] [CrossRef]
- Ji, Y.; Li, X.; Lu, Y.; Guo, P.; Zhang, G.; Wang, Y.; Zhang, Y.; Zhu, W.; Pan, J.; Wang, J. Nanobodies Based on a Sandwich Immunoassay for the Detection of Staphylococcal Enterotoxin B Free from Interference by Protein A. J. Agric. Food Chem. 2020, 68, 5959–5968. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Frenken, L.; Bourgeois, D.; de Ron, L.; Bos, W.; Verrips, T.; Tegonil, M. The crystal structure of a llama heavy chain variable domain. Nat. Struct. Biol. 1996, 3, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steeland, S.; Vandenbroucke, R.E.; Libert, C. Nanobodies as therapeutics: Big opportunities for small antibodies. Drug Discov. Today 2016, 21, 1076–1113. [Google Scholar] [CrossRef]
- Fridy, P.C.; Li, Y.; Keegan, S.; Thompson, M.K.; Nudelman, I.; Scheid, J.F.; Rout, M.P. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 2014, 11, 1253–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Percipalle, M.; Hunashal, Y.; Steyaert, J.; Fogolari, F.; Esposito, G. Structure of nanobody Nb23. Molecules 2021, 26, 3567. [Google Scholar] [CrossRef]
- Desmyter, A.; Spinelli, S.; Roussel, A.; Cambillau, C. Camelid nanobodies: Killing two birds with one stone. Curr. Opin. Struct. Biol. 2015, 32, 1–8. [Google Scholar] [CrossRef]
- Zheng, F.; Zhou, J.; Ouyang, Z.; Zhang, J.; Wang, X.; Muyldermans, S.; Raes, G. Development and characterization of nanobodies targeting the kupffer cell. Front. Immunol. 2021, 12, 391. [Google Scholar] [CrossRef]
- Sun, Z.; Lv, J.; Liu, X.; Tang, Z.; Wang, X.; Xu, Y.; Hammock, B.D. Development of a nanobody-aviTag fusion protein and its application in a streptavidin-biotin-amplified enzyme-linked immunosorbent assay for ochratoxin A in cereal. Anal. Chem. 2018, 90, 10628–10634. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.F.; Yang, Y.Y.; Wan, D.B.; Vasylieva, N.; Zhang, Y.Q.; Hammock, B.D. Chemiluminescent enzyme immunoassay and bioluminescent enzyme immunoassay for tenuazonic acid mycotoxin by exploitation of nanobody and nanobody-nanoluciferase fusion. Anal. Chem. 2020, 92, 11935–11942. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Z.-F.; Wan, D.-B.; Vasylieva, N.; Shen, Y.-D.; Xu, Z.-L.; Yang, J.-Y.; Gettemans, J.; Wang, H.; Hammock, B.D.; et al. Enhanced Non-Toxic Immunodetection of Alternaria Mycotoxin Tenuazonic Acid Based on Ferritin-Displayed Anti-Idiotypic Nanobody-Nanoluciferase Multimers. J. Agric. Food Chem. 2021, 69, 4911–4917. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Lee, Y.; Devaraj, V.; Nguyen, T.M.; Kim, Y.J.; Kim, Y.H.; Oh, J.W. Investigation of colorimetric biosensor array based on programable surface chemistry of M13 bacteriophage towards artificial nose for volatile organic compound detection: From basic properties of the biosensor to practical application. Biosens. Bioelectron. 2021, 188, 113339. [Google Scholar] [CrossRef] [PubMed]
- Blaik, R.A.; Lan, E.; Huang, Y.; Dunn, B. Gold-coated M13 bacteriophage as a template for glucose oxidase biofuel cells with direct electron transfer. ACS Nano 2016, 10, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Xu, Y.; He, Q.; Fu, J.; Liu, X.; Tao, Y. Isolation and characterisation of deoxynivalenol affinity binders from a phage display library based on single-domain camelid heavy chain antibodies (VHHs). Food Agric. Immunol. 2012, 23, 123–131. [Google Scholar] [CrossRef]
- He, Y.; Ren, Y.; Guo, B.; Yang, Y.; Ji, Y.; Zhang, D.; Wang, J.; Wang, Y.; Wang, H. Development of a specific nanobody and its application in rapid and selective determination of Salmonella enteritidis in milk. Food Chem. 2020, 310, 125942. [Google Scholar] [CrossRef]
- Gitlin, A.D.; Shulman, Z.; Nussenzweig, M.C. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 2017, 509, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.C.; Choong, Y.S.; Lim, T.S. Cognizance of molecular methods for the generation of mutagenic phage display antibody libraries for affinity maturation. Int. J. Mol. Sci. 2019, 20, 1861. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Sang, Z.; Bitton, L.; Xu, J.; Liu, Y.; Schneidman-Duhovny, D.; Shi, Y. Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst. 2021, 12, 220–234. [Google Scholar] [CrossRef]
- Bathula, N.V.; Bommadevara, H.; Hayes, J.M. Nanobodies: The Future of Antibody-Based Immune Therapeutics. Cancer Biother. Radiopharm. 2021, 36, 109–122. [Google Scholar] [CrossRef]
- Spero, L.; Morlock, B.A.; Metzger, J.F. On the cross-reactivity of staphylococcal enterotoxins A, B, and C. J. Immunol. 1978, 120, 86–89. [Google Scholar]
- Singh, B.R.; Evenson, M.L.; Bergdoll, M.S. Structural analysis of staphylococcal enterotoxins B and C1 using circular dichroism and fluorescence spectroscopy. Biochemistry 1988, 27, 8735–8741. [Google Scholar] [CrossRef]
- Singh, B.R.; Fu, F.-N.; LeDoux, D.N. Crystal and solution structures of superantigenic staphylococcal enterotoxins compared. Nat. Struct. Mol. Biol. 1994, 1, 358–360. [Google Scholar] [CrossRef]
- Hun, X.; Zhang, Z. A novel sensitive staphylococcal enterotoxin C1 fluoroimmunoassay based on functionalized fluorescent core-shell nanoparticle labels. Food Chem. 2007, 105, 1623–1629. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Z.; Chen, L.; Ma, L. Chemiluminescent imaging detection of staphylococcal enterotoxin C1 in milk and water samples. Food Chem. 2006, 97, 355–360. [Google Scholar] [CrossRef]
- Wang, W.; Liu, L.; Xu, L.; Kuang, H.; Zhu, J.; Xu, C. Gold-Nanoparticle-Based Multiplexed Immunochromatographic Strip for Simultaneous Detection of Staphylococcal Enterotoxin A, B, C, D, and E. Part. Part. Syst. Charact. 2016, 33, 388–395. [Google Scholar] [CrossRef]






| Phage-Displayed Nbs | Capture Nbs (POD450/NOD450) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C2 | C3 | C6 | C7 | C11 | C15 | C19 | C49 | C61 | C64 | C91 | |
| C2 | 1.14 | 2.03 | 1.52 | 1.12 | 2.08 | 1.08 | 1.81 | 2.07 | 1.29 | 1.06 | 0.81 |
| C3 | 1.80 | 1.75 | 3.29 | 1.64 | 1.95 | 1.59 | 3.44 | 2.08 | 2.27 | 1.70 | 0.86 |
| C6 | 1.00 | 1.83 | 1.21 | 0.95 | 1.87 | 0.92 | 1.82 | 1.98 | 1.21 | 1.24 | 0.78 |
| C7 | 1.05 | 4.12 | 1.87 | 1.71 | 3.34 | 1.32 | 1.58 | 3.68 | 1.54 | 1.77 | 1.16 |
| C11 | 2.74 | 2.32 | 8.08 | 3.53 | 2.15 | 3.34 | 5.61 | 2.42 | 4.00 | 4.43 | 0.94 |
| C15 | 1.43 | 4.25 | 2.78 | 1.31 | 4.33 | 1.21 | 2.51 | 4.37 | 1.44 | 1.52 | 0.88 |
| C19 | 0.96 | 3.24 | 1.43 | 0.96 | 2.98 | 1.26 | 2.45 | 2.75 | 1.84 | 1.15 | 0.84 |
| C49 | 1.57 | 6.07 | 4.09 | 1.94 | 5.13 | 1.10 | 2.64 | 4.35 | 2.02 | 2.81 | 1.80 |
| C61 | 1.71 | 2.45 | 5.57 | 3.84 | 2.61 | 3.25 | 3.35 | 2.43 | 3.00 | 5.22 | 0.90 |
| C64 | 1.05 | 2.36 | 1.86 | 1.37 | 2.09 | 1.08 | 1.41 | 2.18 | 1.30 | 1.58 | 0.94 |
| C91 | 1.72 | 7.51 | 5.30 | 2.89 | 6.48 | 1.95 | 3.19 | 7.23 | 2.49 | 3.03 | 0.81 |
| Sample | Spiked Levels (ng/mL) | Detection Level (ng/mL) (ng/mL) (n = 3 a) | Recovery (%) |
|---|---|---|---|
| Fresh milk | 250 | 255.22 ± 13.76 | 102.09 ± 5.51 |
| 500 | 497.42 ± 42.47 | 99.48 ± 8.49 | |
| 1000 | 1000.05 ± 90.34 | 100.05 ± 9.03 | |
| Yogurt in shelf-life | 250 | 233.79 ± 21.88 | 93.52 ± 8.75 |
| 500 | 540.32 ± 36.27 | 108.06 ± 7.25 | |
| 1000 | 950.42 ± 119.42 | 95.04 ± 11.94 | |
| Cheese | 250 | 222.81 ± 9.92 | 89.12 ± 3.97 |
| 500 | 422.62 ± 12.62 | 84.52 ± 2.52 | |
| 1000 | 1020.48 ± 90.03 | 102.05 ± 9.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations All authors have read and agreed to the published version of the manuscript.. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Y.; Chen, L.; Wang, Y.; Zhang, K.; Wu, H.; Liu, Y.; Wang, Y.; Wang, J. Development of a Double Nanobody-Based Sandwich Immunoassay for the Detecting Staphylococcal Enterotoxin C in Dairy Products. Foods 2021, 10, 2426. https://doi.org/10.3390/foods10102426
Ji Y, Chen L, Wang Y, Zhang K, Wu H, Liu Y, Wang Y, Wang J. Development of a Double Nanobody-Based Sandwich Immunoassay for the Detecting Staphylococcal Enterotoxin C in Dairy Products. Foods. 2021; 10(10):2426. https://doi.org/10.3390/foods10102426
Chicago/Turabian StyleJi, Yanwei, Lili Chen, Yingying Wang, Kaihui Zhang, Haofen Wu, Yuan Liu, Yanru Wang, and Jianlong Wang. 2021. "Development of a Double Nanobody-Based Sandwich Immunoassay for the Detecting Staphylococcal Enterotoxin C in Dairy Products" Foods 10, no. 10: 2426. https://doi.org/10.3390/foods10102426