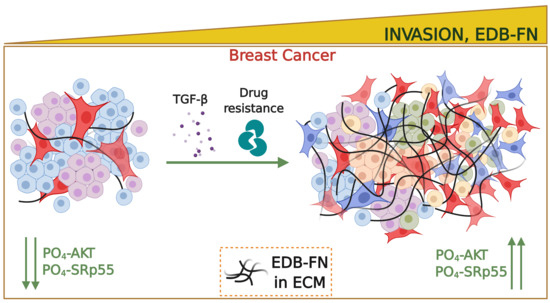
Overexpression of Extradomain-B Fibronectin is Associated with Invasion of Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell lines and Culture
2.2. MK2206-HCl Treatment
2.3. Gene Data Analyses
2.4. Immunohistochemistry of Human Tissue Specimens
2.5. qRT-PCR
2.6. Tumor Spheroid Growth in 3D culture and ZD2-Cy5.5 Staining
2.7. Western Blotting
2.8. Transwell Assay
2.9. Gene Knockdown Assays
2.10. Statistical Analyses
3. Results
3.1. EDB-FN Is Significantly Elevated in Breast Cancer
3.2. Morphological, Functional, and Molecular Changes in 2D- and 3D-Cultured Breast Cancer Cells with TGF-β Treatment and Drug Resistance
3.3. Increased EDB-FN Expression in Breast Cancer Cells with TGF-β Treatment and Drug Resistance
3.4. Therapeutic Ablation of Phospho-AKT in Invasive Breast Cancer Cells Decreases Invasion and EDB-FN Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Goding Sauer, A.; Newman, L.A.; Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Baird, R.D.; Caldas, C. Genetic heterogeneity in breast cancer: The road to personalized medicine? BMC Med. 2013, 11, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Eliyatkin, N.; Yalcin, E.; Zengel, B.; Aktas, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Eccles, S.A.; Aboagye, E.O.; Ali, S.; Anderson, A.S.; Armes, J.; Berditchevski, F.; Blaydes, J.P.; Brennan, K.; Brown, N.J.; Bryant, H.E.; et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013, 15, R92. [Google Scholar] [CrossRef] [Green Version]
- Bleyer, A.; Welch, H.G. Effect of three decades of screening mammography on breast-cancer incidence. N. Engl. J. Med. 2012, 367, 1998–2005. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [Green Version]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci 2002, 115, 3861–3863. [Google Scholar] [CrossRef] [Green Version]
- White, E.S.; Baralle, F.E.; Muro, A.F. New insights into form and function of fibronectin splice variants. J. Pathol. 2008, 216, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Lu, Z.R. Targeting Fibronectin for Cancer Imaging and Therapy. J. Mater. Chem. B 2017, 5, 639–654. [Google Scholar] [CrossRef] [Green Version]
- Loridon-Rosa, B.; Vielh, P.; Matsuura, H.; Clausen, H.; Cuadrado, C.; Burtin, P. Distribution of oncofetal fibronectin in human mammary tumors: Immunofluorescence study on histological sections. Cancer Res. 1990, 50, 1608–1612. [Google Scholar] [PubMed]
- Lyons, A.J.; Bateman, A.C.; Spedding, A.; Primrose, J.N.; Mandel, U. Oncofetal fibronectin and oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 2001, 39, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kim, H.S.; Saw, P.E.; Jon, S.; Moon, W.K. Targeted Therapy for Breast Cancer Stem Cells by Liposomal Delivery of siRNA against Fibronectin EDB. Adv. Healthc. Mater. 2015, 4, 1675–1680. [Google Scholar] [CrossRef] [PubMed]
- Tavian, D.; De Petro, G.; Colombi, M.; Portolani, N.; Giulini, S.M.; Gardella, R.; Barlati, S. RT-PCR detection of fibronectin EDA+ and EDB+ mRNA isoforms: Molecular markers for hepatocellular carcinoma. Int. J. Cancer 1994, 56, 820–825. [Google Scholar] [CrossRef]
- Coltrini, D.; Ronca, R.; Belleri, M.; Zardi, L.; Indraccolo, S.; Scarlato, V.; Giavazzi, R.; Presta, M. Impact of VEGF-dependent tumour micro-environment on EDB fibronectin expression by subcutaneous human tumour xenografts in nude mice. J. Pathol. 2009, 219, 455–462. [Google Scholar] [CrossRef]
- Ventura, E.; Weller, M.; Macnair, W.; Eschbach, K.; Beisel, C.; Cordazzo, C.; Claassen, M.; Zardi, L.; Burghardt, I. TGF-beta induces oncofetal fibronectin that, in turn, modulates TGF-beta superfamily signaling in endothelial cells. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Cheng, H.; Parvani, J.G.; Zhou, Z.; Lu, Z.R. Magnetic resonance molecular imaging of metastatic breast cancer by targeting extradomain-B fibronectin in the tumor microenvironment. Magn. Reson. Med. 2018, 79, 3135–3143. [Google Scholar] [CrossRef]
- Santimaria, M.; Moscatelli, G.; Viale, G.L.; Giovannoni, L.; Neri, G.; Viti, F.; Leprini, A.; Borsi, L.; Castellani, P.; Zardi, L.; et al. Immunoscintigraphic detection of the ED-B domain of fibronectin, a marker of angiogenesis, in patients with cancer. Clin. Cancer Res. 2003, 9, 571–579. [Google Scholar]
- Menzin, A.W.; Loret de Mola, J.R.; Bilker, W.B.; Wheeler, J.E.; Rubin, S.C.; Feinberg, R.F. Identification of oncofetal fibronectin in patients with advanced epithelial ovarian cancer: Detection in ascitic fluid and localization to primary sites and metastatic implants. Cancer 1998, 82, 152–158. [Google Scholar] [CrossRef]
- Giannini, R.; Faviana, P.; Cavinato, T.; Elisei, R.; Pacini, F.; Berti, P.; Fontanini, G.; Ugolini, C.; Camacci, T.; De Ieso, K.; et al. Galectin-3 and oncofetal-fibronectin expression in thyroid neoplasia as assessed by reverse transcription-polymerase chain reaction and immunochemistry in cytologic and pathologic specimens. Thyroid 2003, 13, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhou, Z.; Shi, X.; Wang, J.; Wu, X.; Sun, D.; Chen, Y.; Zhu, H.; Magi-Galluzzi, C.; Lu, Z.R. EDB Fibronectin Specific Peptide for Prostate Cancer Targeting. Bioconjug. Chem. 2015, 26, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Li, Y.; Roelle, S.; Zhou, Z.; Liu, Y.; Sabatelle, R.; DeSanto, A.; Yu, X.; Zhu, H.; Magi-Galluzzi, C.; et al. Targeted Contrast Agent Specific to an Oncoprotein in Tumor Microenvironment with the Potential for Detection and Risk Stratification of Prostate Cancer with MRI. Bioconjug. Chem. 2017, 28, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wu, X.; Roelle, S.; Chen, C.; Schiemann, W.P.; Lu, Z.R. Targeted gadofullerene for sensitive magnetic resonance imaging and risk-stratification of breast cancer. Nat. Commun. 2017, 8, 692. [Google Scholar] [CrossRef] [Green Version]
- Ayat, N.R.; Qin, J.C.; Cheng, H.; Roelle, S.; Gao, S.; Li, Y.; Lu, Z.R. Optimization of ZD2 Peptide Targeted Gd(HP-DO3A) for Detection and Risk-Stratification of Prostate Cancer with MRI. ACS Med. Chem. Lett. 2018, 9, 730–735. [Google Scholar] [CrossRef]
- Sun, Y.; Kim, H.S.; Park, J.; Li, M.; Tian, L.; Choi, Y.; Choi, B.I.; Jon, S.; Moon, W.K. MRI of breast tumor initiating cells using the extra domain-B of fibronectin targeting nanoparticles. Theranostics 2014, 4, 845–857. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.X.; Zhao, Y.Y.; Wang, Q.; Xiao, W.; Zhao, J.; Peng, Y.J.; Cao, D.H.; Lin, W.J.; Si-Tu, M.Y.; Li, M.Z.; et al. EDB Fibronectin-Specific SPECT Probe (99m)Tc-HYNIC-ZD2 for Breast Cancer Detection. ACS Omega 2017, 2, 2459–2468. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Zhang, S.; Fujiwara, K.; Zhang, J.; Li, Y.; Liu, J.; van Zijl, P.C.M.; Lu, Z.R.; Zheng, L.; Liu, G. Extradomain-B Fibronectin-Targeted Dextran-Based Chemical Exchange Saturation Transfer Magnetic Resonance Imaging Probe for Detecting Pancreatic Cancer. Bioconjug. Chem. 2019, 30, 1425–1433. [Google Scholar] [CrossRef]
- Agarwal, E.; Chaudhuri, A.; Leiphrakpam, P.D.; Haferbier, K.L.; Brattain, M.G.; Chowdhury, S. Akt inhibitor MK-2206 promotes anti-tumor activity and cell death by modulation of AIF and Ezrin in colorectal cancer. BMC Cancer 2014, 14, 145. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, A.M.; Sun, Z.; Ayat, N.; Schilb, A.; Liu, X.; Jiang, H.; Sun, D.; Scheidt, J.; Qian, V.; He, S.; et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjug. Chem. 2019, 30, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Gujrati, M.; Vaidya, A.M.; Mack, M.; Snyder, D.; Malamas, A.; Lu, Z.R. Targeted Dual pH-Sensitive Lipid ECO/siRNA Self-Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Adv. Healthc. Mater. 2016, 5, 2882–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnedos, M.; Bihan, C.; Delaloge, S.; Andre, F. Triple-negative breast cancer: Are we making headway at least? Adv. Med. Oncol. 2012, 4, 195–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, C.; Zhang, J.; Xu, W.; Zhang, Y. Progress of CDK4/6 Inhibitor Palbociclib in the Treatment of Cancer. Anticancer Agents Med. Chem. 2018, 18, 1241–1251. [Google Scholar] [CrossRef]
- Barbuti, A.M.; Chen, Z.S. Paclitaxel Through the Ages of Anticancer Therapy: Exploring Its Role in Chemoresistance and Radiation Therapy. Cancers (Basel) 2015, 7, 2360–2371. [Google Scholar] [CrossRef]
- Fedele, M.; Cerchia, L.; Chiappetta, G. The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas. Cancers (Basel) 2017, 9, 134. [Google Scholar] [CrossRef]
- Huang, J.; Li, H.; Ren, G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review). Int. J. Oncol. 2015, 47, 840–848. [Google Scholar] [CrossRef] [Green Version]
- Kroger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, H.; Sootome, H.; Nakatsuru, Y.; Miyama, K.; Taguchi, S.; Tsujioka, K.; Ueno, Y.; Hatch, H.; Majumder, P.K.; Pan, B.S.; et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer 2010, 9, 1956–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordeleau, F.; Califano, J.P.; Negron Abril, Y.L.; Mason, B.N.; LaValley, D.J.; Shin, S.J.; Weiss, R.S.; Reinhart-King, C.A. Tissue stiffness regulates serine/arginine-rich protein-mediated splicing of the extra domain B-fibronectin isoform in tumors. Proc. Natl. Acad. Sci. USA 2015, 112, 8314–8319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatcheressian, J.L.; Hurley, P.; Bantug, E.; Esserman, L.J.; Grunfeld, E.; Halberg, F.; Hantel, A.; Henry, N.L.; Muss, H.B.; Smith, T.J.; et al. Breast cancer follow-up and management after primary treatment: American Society of Clinical Oncology clinical practice guideline update. J. Clin. Oncol. 2013, 31, 961–965. [Google Scholar] [CrossRef]
- Bayo, J.; Castano, M.A.; Rivera, F.; Navarro, F. Analysis of blood markers for early breast cancer diagnosis. Clin. Transl. Oncol. 2018, 20, 467–475. [Google Scholar] [CrossRef]
- Othman, E.; Wang, J.; Sprague, B.L.; Rounds, T.; Ji, Y.; Herschorn, S.D.; Wood, M.E. Comparison of false positive rates for screening breast magnetic resonance imaging (MRI) in high risk women performed on stacked versus alternating schedules. Springerplus 2015, 4, 77. [Google Scholar] [CrossRef] [Green Version]
- Delou, J.M.A.; Souza, A.S.O.; Souza, L.C.M.; Borges, H.L. Highlights in Resistance Mechanism Pathways for Combination Therapy. Cells 2019, 8, 1013. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.C.; Yang, C.H.; Cheng, L.H.; Chang, W.T.; Lin, Y.R.; Cheng, H.C. Fibronectin in Cancer: Friend or Foe. Cells 2019, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, E.; Yoshida, T.; Kawarada, Y.; Sakakura, T. Expression of fibronectin isoforms in human breast tissue: Production of extra domain A+/extra domain B+ by cancer cells and extra domain A+ by stromal cells. Jpn. J. Cancer Res. 1999, 90, 320–325. [Google Scholar] [CrossRef]
- Kwon, A.; Chae, I.H.; You, E.; Kim, S.H.; Ahn, S.Y.; Lee, O.J.; Park, Z.Y.; Rhee, S.; Huh, Y.H.; Song, W.K. Extra domain A-containing fibronectin expression in Spin90-deficient fibroblasts mediates cancer-stroma interaction and promotes breast cancer progression. J. Cell Physiol. 2020, 235, 4494–4507. [Google Scholar] [CrossRef]
- Huh, G.S.; Hynes, R.O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994, 8, 1561–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ghigna, C.; Valacca, C.; Biamonti, G. Alternative splicing and tumor progression. Curr. Genom. 2008, 9, 556–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stottrup, C.; Tsang, T.; Chin, Y.R. Upregulation of AKT3 Confers Resistance to the AKT Inhibitor MK2206 in Breast Cancer. Mol. Cancer 2016, 15, 1964–1974. [Google Scholar] [CrossRef] [Green Version]
- Arnold, S.A.; Loomans, H.A.; Ketova, T.; Andl, C.D.; Clark, P.E.; Zijlstra, A. Urinary oncofetal ED-A fibronectin correlates with poor prognosis in patients with bladder cancer. Clin. Exp. Metastasis 2016, 33, 29–44. [Google Scholar] [CrossRef] [Green Version]
- Khan, Z.A.; Caurtero, J.; Barbin, Y.P.; Chan, B.M.; Uniyal, S.; Chakrabarti, S. ED-B fibronectin in non-small cell lung carcinoma. Exp. Lung Res. 2005, 31, 701–711. [Google Scholar] [CrossRef]
- Albrecht, M.; Renneberg, H.; Wennemuth, G.; Moschler, O.; Janssen, M.; Aumuller, G.; Konrad, L. Fibronectin in human prostatic cells in vivo and in vitro: Expression, distribution, and pathological significance. Histochem. Cell Biol. 1999, 112, 51–61. [Google Scholar] [CrossRef]
- Mariani, G.; Lasku, A.; Balza, E.; Gaggero, B.; Motta, C.; Di Luca, L.; Dorcaratto, A.; Viale, G.A.; Neri, D.; Zardi, L. Tumor targeting potential of the monoclonal antibody BC-1 against oncofetal fibronectin in nude mice bearing human tumor implants. Cancer 1997, 80, 2378–2384. [Google Scholar] [CrossRef]
- Tijink, B.M.; Perk, L.R.; Budde, M.; Stigter-van Walsum, M.; Visser, G.W.; Kloet, R.W.; Dinkelborg, L.M.; Leemans, C.R.; Neri, D.; van Dongen, G.A. (124)I-L19-SIP for immuno-PET imaging of tumour vasculature and guidance of (131)I-L19-SIP radioimmunotherapy. Eur. J. Nucl. Med. Mol. Imaging 2009, 36, 1235–1244. [Google Scholar] [CrossRef] [Green Version]
- Zahnd, C.; Kawe, M.; Stumpp, M.T.; de Pasquale, C.; Tamaskovic, R.; Nagy-Davidescu, G.; Dreier, B.; Schibli, R.; Binz, H.K.; Waibel, R.; et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: Effects of affinity and molecular size. Cancer Res. 2010, 70, 1595–1605. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaidya, A.; Wang, H.; Qian, V.; Gilmore, H.; Lu, Z.-R. Overexpression of Extradomain-B Fibronectin is Associated with Invasion of Breast Cancer Cells. Cells 2020, 9, 1826. https://doi.org/10.3390/cells9081826
Vaidya A, Wang H, Qian V, Gilmore H, Lu Z-R. Overexpression of Extradomain-B Fibronectin is Associated with Invasion of Breast Cancer Cells. Cells. 2020; 9(8):1826. https://doi.org/10.3390/cells9081826
Chicago/Turabian StyleVaidya, Amita, Helen Wang, Victoria Qian, Hannah Gilmore, and Zheng-Rong Lu. 2020. "Overexpression of Extradomain-B Fibronectin is Associated with Invasion of Breast Cancer Cells" Cells 9, no. 8: 1826. https://doi.org/10.3390/cells9081826