WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line
Abstract
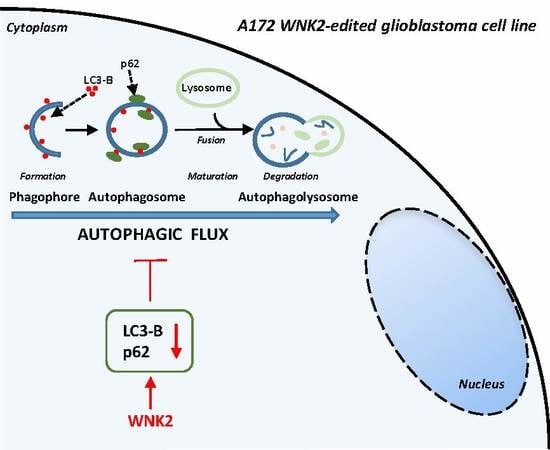
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Cell Treatment
2.3. Immunofluorescence
2.4. Transient Transfection
2.5. Western Blotting Analysis
2.6. Statistical Analysis
3. Results
WNK2 Inhibits Autophagic Flux
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro-Oncology 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Parada, L.F. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer 2002, 2, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Cruceru, M.L.; Neagu, M.; Demoulin, J.B.; Constantinescu, S.N. Therapy targets in glioblastoma and ncancer stem cells: Lessons from haematopoietic neoplasms. J. Cell. Mol. Med. 2013, 17, 1218–1235. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef]
- Yuanchao, S.; Xunsi, Q.; Hong, C.; Wei, S. Epigenetic control of autophagy. Yi Chuan Hered. 2014, 36, 447–455. [Google Scholar]
- Yan, Y.; Xu, Z.; Dai, S.; Qian, L.; Sun, L.; Gong, Z. Targeting autophagy to sensitive glioma to temozolomide treatment. J. Exp. Clin. Cancer Res. 2016, 35, 23. [Google Scholar] [CrossRef] [Green Version]
- Eskelinen, E.L. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011, 11, 294–300. [Google Scholar] [CrossRef]
- Notte, A.; Leclere, L.; Michiels, C. Autophagy as a mediator of chemotherapy-induced cell death in cancer. Biochem. Pharmacol. 2011, 82, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Levine, B. Autophagic cell death: The story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; White, E.J.; Conrad, C.; Gomez-Manzano, C.; Fueyo, J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009, 453, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Golden, E.B.; Cho, H.Y.; Jahanian, A.; Hofman, F.M.; Louie, S.G.; Schonthal, A.H.; Chen, T.C. Chloroquine enhances temozolomide cytotoxicity in malignant gliomas by blocking autophagy. Neurosurg. Focus 2014, 37, E12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.; Moorefield, K.S.; Jun, P.; Aldape, K.D.; Kharbanda, S.; Phillips, H.S.; Costello, J.F. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc. Natl. Acad. Sci. USA 2007, 104, 10974–10979. [Google Scholar] [CrossRef] [Green Version]
- Jun, P.; Hong, C.; Lal, A.; Wong, J.M.; McDermott, M.W.; Bollen, A.W.; Plass, C.; Held, W.A.; Smiraglia, D.J.; Costello, J.F. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro-Oncology 2009, 11, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Moniz, S.; Martinho, O.; Pinto, F.; Sousa, B.; Loureiro, C.; Oliveira, M.J.; Moita, L.F.; Honavar, M.; Pinheiro, C.; Pires, M.; et al. Loss of WNK2 expression by promoter gene methylation occurs in adult gliomas and triggers Rac1-mediated tumour cell invasiveness. Hum. Mol. Genet. 2013, 22, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Moniz, S.; Verissimo, F.; Matos, P.; Brazao, R.; Silva, E.; Kotelevets, L.; Chastre, E.; Gespach, C.; Jordan, P. Protein kinase WNK2 inhibits cell proliferation by negatively modulating the activation of MEK1/ERK1/2. Oncogene 2007, 26, 6071–6081. [Google Scholar] [CrossRef] [Green Version]
- Moniz, S.; Matos, P.; Jordan, P. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cell. Signal. 2008, 20, 1762–1768. [Google Scholar] [CrossRef]
- Costa, A.M.; Pinto, F.; Martinho, O.; Oliveira, M.J.; Jordan, P.; Reis, R.M. Silencing of the tumor suppressor gene WNK2 is associated with upregulation of MMP2 and JNK in gliomas. Oncotarget 2015, 6, 1422–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Liang, Y.; Murphy, S.F.; Huang, A.; Shen, H.; Kelly, D.F.; Sobrado, P.; Sheng, Z. A rapid and high content assay that measures cyto-ID-stained autophagic compartments and estimates autophagy flux with potential clinical applications. Autophagy 2015, 11, 560–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szyniarowski, P.; Corcelle-Termeau, E.; Farkas, T.; Hoyer-Hansen, M.; Nylandsted, J.; Kallunki, T.; Jaattela, M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011, 7, 892–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Oliveira, R.J.; Silva, V.A.; Martinho, O.; Cruvinel-Carloni, A.; Melendez, M.E.; Rosa, M.N.; de Paula, F.E.; de Souza Viana, L.; Carvalho, A.L.; Reis, R.M. Cytotoxicity of allitinib, an irreversible anti-EGFR agent, in a large panel of human cancer-derived cell lines: KRAS mutation status as a predictive biomarker. Cell. Oncol. 2016, 39, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, T.L.; Oliveira Silva, V.A.; da Cunha, D.B.; Polettini, F.L.; Thomaz, C.D.; Pianca, A.A.; Zambom, F.L.; da Silva Leitao Mazzi, D.P.; Reis, R.M.; Mazzi, M.V. Isolation, characterization and screening of the in vitro cytotoxic activity of a novel L-amino acid oxidase (LAAOcdt) from Crotalus durissus terrificus venom on human cancer cell lines. Toxicon 2016, 119, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Bravo-San Pedro, J.M.; Niso-Santano, M.; Gomez-Sanchez, R.; Pizarro-Estrella, E.; Aiastui-Pujana, A.; Gorostidi, A.; Climent, V.; Lopez de Maturana, R.; Sanchez-Pernaute, R.; Lopez de Munain, A.; et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell. Mol. Life Sci. 2013, 70, 121–136. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Furnari, F.B.; Fenton, T.; Bachoo, R.M.; Mukasa, A.; Stommel, J.M.; Stegh, A.; Hahn, W.C.; Ligon, K.L.; Louis, D.N.; Brennan, C.; et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007, 21, 2683–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Daido, S.; Kanzawa, T.; Kondo, S.; Kondo, Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int. J. Oncol. 2005, 26, 1401–1410. [Google Scholar] [CrossRef]
- Iwamaru, A.; Kondo, Y.; Iwado, E.; Aoki, H.; Fujiwara, K.; Yokoyama, T.; Mills, G.B.; Kondo, S. Silencing mammalian target of rapamycin signaling by small interfering RNA enhances rapamycin-induced autophagy in malignant glioma cells. Oncogene 2007, 26, 1840–1851. [Google Scholar] [CrossRef] [Green Version]
- Murugan, A.K. mTOR: Role in cancer, metastasis and drug resistance. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838. [Google Scholar] [CrossRef]
- Li, S.; Jiang, T.; Li, G.; Wang, Z. Impact of p53 status to response of temozolomide in low MGMT expression glioblastomas: Preliminary results. Neurol. Res. 2008, 30, 567–570. [Google Scholar] [CrossRef]
- Carmo, A.; Carvalheiro, H.; Crespo, I.; Nunes, I.; Lopes, M.C. Effect of temozolomide on the U-118 glioma cell line. Oncol. Lett. 2011, 2, 1165–1170. [Google Scholar] [CrossRef] [Green Version]
- Kanzawa, T.; Germano, I.M.; Komata, T.; Ito, H.; Kondo, Y.; Kondo, S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004, 11, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Filippi-Chiela, E.C.; Bueno e Silva, M.M.; Thome, M.P.; Lenz, G. Single-cell analysis challenges the connection between autophagy and senescence induced by DNA damage. Autophagy 2015, 11, 1099–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenfeld, M.R.; Ye, X.; Supko, J.G.; Desideri, S.; Grossman, S.A.; Brem, S.; Mikkelson, T.; Wang, D.; Chang, Y.C.; Hu, J.; et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014, 10, 1359–1368. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, A.L.V.; Costa, A.M.; Martinho, O.; da Silva, V.D.; Jordan, P.; Silva, V.A.O.; Reis, R.M. WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line. Cells 2020, 9, 485. https://doi.org/10.3390/cells9020485
Alves ALV, Costa AM, Martinho O, da Silva VD, Jordan P, Silva VAO, Reis RM. WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line. Cells. 2020; 9(2):485. https://doi.org/10.3390/cells9020485
Chicago/Turabian StyleAlves, Ana Laura Vieira, Angela Margarida Costa, Olga Martinho, Vinicius Duval da Silva, Peter Jordan, Viviane Aline Oliveira Silva, and Rui Manuel Reis. 2020. "WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line" Cells 9, no. 2: 485. https://doi.org/10.3390/cells9020485