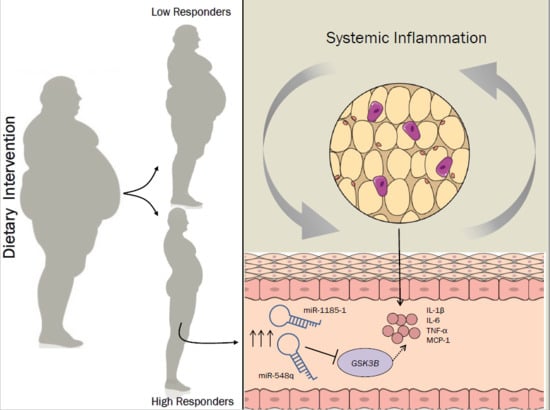
miR-1185-1 and miR-548q Are Biomarkers of Response to Weight Loss and Regulate the Expression of GSK3B
Abstract
:1. Introduction
2. Experimental Section
2.1. RNA Isolation and Reverse Transcription
2.2. Microarray Analyses and miRNA-Seq
2.3. Bioinformatic Study
2.4. Luciferase Reporter Constructs
2.5. Cell Culture
mirVana miRNA Mimic Transfections
2.6. Dual-Luciferase Reporter Assays
2.7. Quantitative Real-Time PCR
2.8. Statistical Analysis
3. Results
3.1. Dietary Intervention
3.2. miR-548q and miR-1185-1 Are Overexpressed in High Responders to the Weight Loss Intervention
3.3. GSK3B Is a Putative Target Gene for miR-548q and miR-1185-1
3.4. miR-1185-1 Binds to the 3′-UTR of GSK3B
3.5. miR-548q and miR-1185-1 Decrease the Endogenous GSK3B mRNA Levels
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; Naghavi, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Goni, L.; Cuervo, M.; Milagro, F.I.; Martinez, J.A. Future Perspectives of Personalized Weight Loss Interventions Based on Nutrigenetic, Epigenetic, and Metagenomic Data. J. Nutr. 2016, 905S–912S. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.A.; Davis, C.D. The emerging role of microRNAs and nutrition in modulating health and disease. Annu. Rev. Nutr. 2014, 34, 305–336. [Google Scholar] [CrossRef]
- Pomatto, M.A.C.; Gai, C.; Deregibus, M.C.; Tetta, C.; Camussi, G. Noncoding RNAs Carried by Extracellular Vesicles in Endocrine Diseases. Int. J. Endocrinol. 2018, 2018, 1–18. [Google Scholar] [CrossRef]
- Houshmand-Oeregaard, A.; Schrolkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.C.B.; Broholm, C.; Mathiesen, E.R.; Clausen, T.D.; Vaag, A.; et al. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martinez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. Faseb. J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef]
- Yu, J.; Lv, Y.; Di, W.; Liu, J.; Kong, X.; Sheng, Y.; Huang, M.; Lv, S.; Qi, H.; Gao, M.; et al. MiR-27b-3p Regulation in Browning of Human Visceral Adipose Related to Central Obesity. Obesity 2018, 26, 387–396. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, L.H.; Su, D.J.; Zhu, D.; Li, Q.M.; Chi, M.H. MicroRNA-29b promotes the adipogenic differentiation of human adipose tissue-derived stromal cells. Obesity 2016, 24, 1097–1105. [Google Scholar] [CrossRef]
- Liu, W.; Cao, H.; Ye, C.; Chang, C.; Lu, M.; Jing, Y.; Zhang, D.; Yao, X.; Duan, Z.; Xia, H.; et al. Hepatic miR-378 targets p110alpha and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat. Commun. 2014, 5, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Wu, W.; Yin, B.; Liu, X.; Ren, F. MicroRNA-463-3p/ABCG4: A new axis in glucose-stimulated insulin secretion. Obesity 2016, 24, 2368–2376. [Google Scholar] [CrossRef] [PubMed]
- Derghal, A.; Djelloul, M.; Airault, C.; Pierre, C.; Dallaporta, M.; Troadec, J.D.; Tillement, V.; Tardivel, C.; Bariohay, B.; Trouslard, J.; et al. Leptin is required for hypothalamic regulation of miRNAs targeting POMC 3’UTR. Front. Cell Neurosci. 2015, 9, 172–193. [Google Scholar] [CrossRef] [PubMed]
- Pescador, N.; Perez-Barba, M.; Ibarra, J.M.; Corbaton, A.; Martinez-Larrad, M.T.; Serrano-Rios, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. miRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Wu, X.; Jiang, L.; Yang, S.; Ding, Z.; Fang, Z.; Hua, H.; Kirby, M.S.; Shou, J. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. BMC Genom. 2018, 19, 188–209. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Roy, D.; Bhattacharyya, M.; Bandyopadhyay, S. Biological networks in Parkinson’s disease: An insight into the epigenetic mechanisms associated with this disease. BMC Genom. 2017, 18, 721–751. [Google Scholar] [CrossRef]
- Landrier, J.F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef]
- Parr, E.B.; Camera, D.M.; Burke, L.M.; Phillips, S.M.; Coffey, V.G.; Hawley, J.A. Circulating MicroRNA Responses between ‘High’ and ‘Low’ Responders to a 16-Wk Diet and Exercise Weight Loss Intervention. PLoS ONE 2016, 11, e0152545. [Google Scholar] [CrossRef]
- Milagro, F.I.; Miranda, J.; Portillo, M.P.; Fernandez-Quintela, A.; Campión, J.; Martínez, J.A. High-Throughput Sequencing of microRNAs in Peripheral Blood Mononuclear Cells: Identification of Potential Weight Loss Biomarkers. PLoS ONE 2013, 8, e54319. [Google Scholar] [CrossRef]
- Garcia-Lacarte, M.; Martinez, J.A.; Zulet, M.A.; Milagro, F.I. Implication of miR-612 and miR-1976 in the regulation of TP53 and CD40 and their relationship in the response to specific weight-loss diets. PLoS ONE 2018, 13, e0201217. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids. Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Wu, Y.; Zhang, X.; Liao, Y.; Sibanda, V.L.; Liu, W.; Guo, A.-Y. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. Rna. Biol. 2014, 11, 1375–1385. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Maury, E.; Brichard, S.M. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol. Cell Endocrinol. 2010, 314, 1–16. [Google Scholar] [CrossRef]
- Varma, V.; Yao-Borengasser, A.; Rasouli, N.; Nolen, G.T.; Phanavanh, B.; Starks, T.; Gurley, C.; Simpson, P.; McGehee, R.E., Jr.; Kern, P.A.; et al. Muscle inflammatory response and insulin resistance: Synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1300–E1310. [Google Scholar] [CrossRef]
- Wang, X.; Ge, A.; Cheng, M.; Guo, F.; Zhao, M.; Zhou, X.; Liu, L.; Yang, N. Increased hypothalamic inflammation associated with the susceptibility to obesity in rats exposed to high-fat diet. Exp. Diabetes Res. 2012, 2012, 847246–847254. [Google Scholar] [CrossRef]
- Chang, R.C.; Ying, W.; Bazer, F.W.; Zhou, B. MicroRNAs Control Macrophage Formation and Activation: The Inflammatory Link between Obesity and Cardiovascular Diseases. Cells 2014, 3, 702–712. [Google Scholar] [CrossRef]
- Feng, J.; Li, A.; Deng, J.; Yang, Y.; Dang, L.; Ye, Y.; Li, Y.; Zhang, W. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: Potential role in cerebrovascular disease. Lipids Health Dis. 2014, 13, 27–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fordham, J.B.; Naqvi, A.R.; Nares, S. miR-24 Regulates Macrophage Polarization and Plasticity. J. Clin. Cell Immunol. 2015, 6, 362–381. [Google Scholar] [PubMed] [Green Version]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jope, R.S.; Yuskaitis, C.J.; Beurel, E. Glycogen Synthase Kinase-3 (GSK3): Inflammation, Diseases, and Therapeutics. Neurochem. Res. 2007, 32, 577–595. [Google Scholar] [CrossRef] [Green Version]
- Steinbrecher, K.A.; Wilson, W., 3rd; Cogswell, P.C.; Baldwin, A.S. Glycogen synthase kinase 3beta functions to specify gene-specific, NF-kappaB-dependent transcription. Mol. Cell. Biol. 2005, 25, 8444–8455. [Google Scholar] [CrossRef] [Green Version]
- Beurel, E. Regulation by Glycogen Synthase Kinase-3 of Inflammation and T Cells in CNS Diseases. Front. Mol. Neurosci. 2011, 4, 18–42. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.; Rehani, K.; Jope, R.S.; Michalek, S.M. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 2005, 6, 777–784. [Google Scholar] [CrossRef]
- Eldar-Finkelman, H.; Schreyer, S.A.; Shinohara, M.M.; LeBoeuf, R.C.; Krebs, E.G. Increased glycogen synthase kinase-3 activity in diabetes- and obesity-prone C57BL/6J mice. Diabetes 1999, 48, 1662–1666. [Google Scholar] [CrossRef]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, Y.; Meng, Y.; Zhang, C.; Di, L. GSK3-activated STAT5 regulates expression of SFRPs to modulate adipogenesis. Faseb. J. 2018, 32, 4714–4726. [Google Scholar] [CrossRef] [Green Version]
- Patricia, S.-B.; Wolfgang, W.; Ralf, K. Simultaneous Cre-mediated conditional knockdown of two genes in mice. Genesis 2008, 46, 144–151. [Google Scholar]
- Pearce, N.J.; Arch, J.R.S.; Clapham, J.C.; Coghlan, M.P.; Corcoran, S.L.; Lister, C.A.; Llano, A.; Moore, G.B.; Murphy, G.J.; Smith, S.A.; et al. Development of glucose intolerance in male transgenic mice overexpressing human glycogen synthase kinase-3β on a muscle-specific promoter. Metabolism 2004, 53, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Yang, W.K.; Song, J.H.; Ra, Y.M.; Jeong, J.-H.; Choe, W.; Kang, I.; Kim, S.-S.; Ha, J. Anti-obesity effects of 3-hydroxychromone derivative, a novel small-molecule inhibitor of glycogen synthase kinase-3. Biochem. Pharm. 2013, 85, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, C.; Esteller, M. Chapter Seven-MicroRNAs and Epigenetics. In Advances in Cancer Research; Croce, C.M., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 135, pp. 189–220. [Google Scholar]
- Tang, X.; Zheng, D.; Hu, P.; Zeng, Z.; Li, M.; Tucker, L.; Monahan, R.; Resnick, M.B.; Liu, M.; Ramratnam, B. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic. Acids Res. 2014, 42, 2988–2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, C.; Wang, R.; Li, D.; Lin, X.J.; Wei, Q.K.; Yuan, Y.; Wang, Q.; Chen, W.; Zhuang, S.M. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology 2010, 52, 1702–1712. [Google Scholar] [CrossRef]
- Ji, J.; Yamashita, T.; Wang, X.W. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011, 1, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. |




| GSK3B-miR-548q-F | 5′-TTAGCTAGCACAGTAGGTACCGGCCTGTA-3′ | 668 bp |
| GSK3B-miR-548q -R | 5′-TTATCTAGAGGTGGCACTCCGTGCAGT-3′ | |
| GSK3B-miR-1185-1-F | 5′-TTTGCTAGCCCGATGGATCACTTGGGCCT-3′ | 856 bp |
| GSK3B-miR-1185-1-R | 5′-TTATCTAGAGGAGGTACAGCCCCACTGTT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Lacarte, M.; Mansego, M.L.; Zulet, M.A.; Martinez, J.A.; Milagro, F.I. miR-1185-1 and miR-548q Are Biomarkers of Response to Weight Loss and Regulate the Expression of GSK3B. Cells 2019, 8, 1548. https://doi.org/10.3390/cells8121548
Garcia-Lacarte M, Mansego ML, Zulet MA, Martinez JA, Milagro FI. miR-1185-1 and miR-548q Are Biomarkers of Response to Weight Loss and Regulate the Expression of GSK3B. Cells. 2019; 8(12):1548. https://doi.org/10.3390/cells8121548
Chicago/Turabian StyleGarcia-Lacarte, Marcos, Maria L. Mansego, M. Angeles Zulet, J. Alfredo Martinez, and Fermin I. Milagro. 2019. "miR-1185-1 and miR-548q Are Biomarkers of Response to Weight Loss and Regulate the Expression of GSK3B" Cells 8, no. 12: 1548. https://doi.org/10.3390/cells8121548