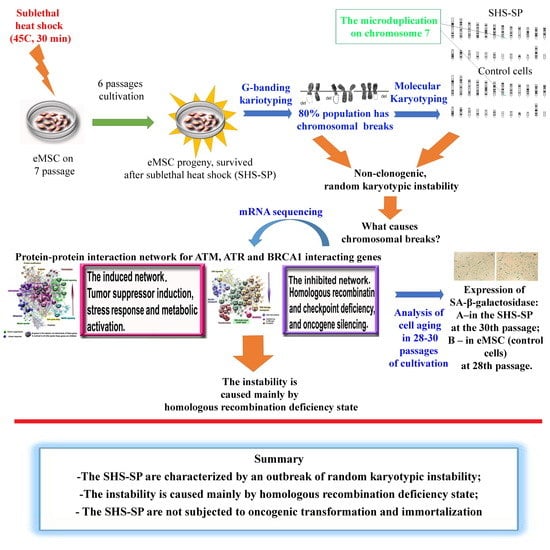
Cytogenetic and Transcriptomic Analysis of Human Endometrial MSC Retaining Proliferative Activity after Sublethal Heat Shock
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. Immunophenotyping
2.3. Adipogenic Differentiation
2.4. Osteogenic Differentiation
2.5. Sublethal Heat Shock (SHS)
2.6. Karyotype Analysis
2.7. Molecular Karyotyping
2.8. mRNA Expression Analysis by Next-Generation Sequencing
2.9. Gene Module Analysis
2.10. Protein-Protein Interaction Network Construction and Analysis
2.11. PPI Network and Cluster Analysis
2.12. The Detection of SA-Β-Galactosidase Activity
3. Results
3.1. Characteristics of eMSC
3.2. eMSC Differentiation In Vitro
3.3. Karyotyping
3.3.1. G-Banded Karyotype of Normal eMSC
3.3.2. G-Banding of SHS-SP
3.3.3. Molecular Karyotyping
3.4. Transcriptome Analysis
3.4.1. Functional Enrichment Analysis of DNA-Repair-Related Gene Modules and Evaluation of Expression of Master DNA Repair Regulators
3.4.2. PPI Network Analysis for ATM, ATR and BRCA1 Interacting Genes
3.4.3. Characterizing of HR and DNA Repair Regulator Thermostability
3.4.4. PPI Network Analysis of Oncogenes and Thermosensitive DNA-Repair-Related Genes
3.4.5. Comparative Analysis of Cancer-Related Gene Set Expression in SHS-SP VS. Unheated Cells and in hTERT Transformed Human Bone Marrow MSC VS. Control
3.5. SHS-SP Replicative Aging Evaluation Using SA-β-gal Activity Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rammal, H.; Harmouch, C.; Lataillade, J.J.; Laurent-Maquin, D.; Labrude, P.; Menu, P.; Kerdjoudj, H. Stem cells: A promising source for vascular regenerative medicine. Stem Cells Dev. 2014, 23, 2931–2949. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.V.; Zatsepina, O.G.; Garbuz, D.G.; Evgen’ev, M.B.; Belyavsky, A.V. Recombinant HSP70 and mild heat shock stimulate growth of aged mesenchymal stem cells. Cell Stress Chaperones 2016, 21, 727–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Meng, F.; Cox, C.S.; Lally, K.P.; Huard, J.; Li, Y. Regeneration and Regrowth Potentials of Digit Tips in Amphibians and Mammals. Int. J. Cell Biol. 2017, 2017, 5312951. [Google Scholar] [CrossRef] [PubMed]
- Gornostaeva, A.N.; Andreeva, E.R.; Bobyleva, P.I.; Buravkova, L.B. Interaction of allogeneic adipose tissue-derived stromal cells and unstimulated immune cells in vitro: The impact of cell-to-cell contact and hypoxia in the local milieu. Cytotechnology 2018, 70, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Buravkova, L.B.; Rudimov, E.G.; Andreeva, E.R.; Grigoriev, A.I. The ICAM-1 expression level determines the susceptibility of human endothelial cells to simulated microgravity. J. Cell. Biochem. 2018, 119, 2875–2885. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells-current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef] [PubMed]
- Vojnits, K.; Pan, H.; Dai, X.; Sun, H.; Tong, Q.; Darabi, R.; Huard, J.; Li, Y. Functional neuronal differentiation of injury-induced muscle-derived stem cell-like cells with therapeutic implications. Sci. Rep. 2017, 7, 1177. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.C.; Lippert, T.; Nguyen, H.; Kaelber, S.; Sanberg, P.R.; Borlongan, C.V. Menstrual blood-derived stem cells: In vitro and in vivo characterization of functional effects. Adv. Exp. Med. Biol. 2016, 951, 111–121. [Google Scholar] [PubMed]
- Gurung, S.; Deane, J.; Masuda, H.; Maruyama, T.; Gargett, C. Stem Cells in Endometrial Physiology. Semin. Reprod. Med. 2015, 33, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Gargett, C.E.; Masuda, H. Adult stem cells in the endometrium. Mol. Hum. Reprod. 2010, 16, 818–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Ichim, T.E.; Zhong, J.; Rogers, A.; Yin, Z.; Jackson, J.; Wang, H.; Ge, W.; Bogin, V.; Chan, K.W.; et al. Endometrial regenerative cells: A novel stem cell population. J. Transl. Med. 2007, 5, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musina, R.A.; Belyavski, A.V.; Tarusova, O.V.; Solovyova, E.V.; Sukhikh, G.T. Endometrial mesenchymal stem cells isolated from the menstrual blood. Bull. Exp. Biol. Med. 2008, 145, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.N.; Park, E.; Kuzman, M.; Benetti, F.; Silva, F.J.; Allickson, J.G. Multipotent menstrual blood stromal stem cells: Isolation, characterization, and differentiation. Cell Transplant. 2008, 17, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Verdi, J.; Tan, A.; Shoae-Hassani, A.; Seifalian, A.M. Endometrial stem cells in regenerative medicine. J. Biol. Eng. 2014, 8, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cousins, F.L.; O, D.F.; Gargett, C.E. Endometrial stem/progenitor cells and their role in the pathogenesis of endometriosis. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 50, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Bockeria, L.; Bogin, V.; Bockeria, O.; Le, T.; Alekyan, B.; Woods, E.J.; Brown, A.A.; Ichim, T.E.; Patel, A.N. Endometrial regenerative cells for treatment of heart failure: A new stem cell enters the clinic. J. Transl. Med. 2013, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Ghobadi, A.F.; Elliott, J.R. A simple extrapolation of thermodynamic perturbation theory to infinite order. J. Chem. Phys. 2015, 143, 114107. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, G. Circulating mesenchymal stem cells and their clinical implications. J. Orthop. Transl. 2014, 2, 1–7. [Google Scholar] [CrossRef]
- Li, C.Y.; Wu, X.Y.; Tong, J.B.; Yang, X.X.; Zhao, J.L.; Zheng, Q.F.; Zhao, G.B.; Ma, Z.J. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Res. Ther. 2015, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Staff, N.P.; Madigan, N.N.; Morris, J.; Jentoft, M.; Sorenson, E.J.; Butler, G.; Gastineau, D.; Dietz, A.; Windebank, A.J. Safety of intrathecal autologous adipose-derived mesenchymal stromal cells in patients with ALS. Neurology 2016, 87, 2230–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.Y.; Chiang, C.H.; Hung, S.C.; Chian, C.F.; Tsai, C.L.; Chen, W.C.; Zhang, H. Hypoxia-preconditioned mesenchymal stem cells ameliorate ischemia/reperfusion-induced lung injury. PLoS ONE 2017, 12, e0187637. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.R.; Abadeh, A.; Connelly, K.A. Concise review: Rational use of mesenchymal stem cells in the treatment of ischemic heart disease. Stem Cells Transl. Med. 2018, 7, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Qiao, P.F.; Yao, L.; Zhang, X.C.; Li, G.D.; Wu, D.Q. Heat shock pretreatment improves stem cell repair following ischemia-reperfusion injury via autophagy. World J. Gastroenterol. 2015, 21, 12822–12834. [Google Scholar] [CrossRef] [PubMed]
- Toma, C.; Pittenger, M.F.; Cahill, K.S.; Byrne, B.J.; Kessler, P.D. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation 2002, 105, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Alekseenko, L.L.; Zemelko, V.I.; Zenin, V.V.; Pugovkina, N.A.; Kozhukharova, I.V.; Kovaleva, Z.V.; Grinchuk, T.M.; Fridlyanskaya, I.I.; Nikolsky, N.N. Heat shock induces apoptosis in human embryonic stem cells but a premature senescence phenotype in their differentiated progeny. Cell Cycle 2012, 11, 3260–3269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alekseenko, L.L.; Zemelko, V.I.; Domnina, A.P.; Lyublinskaya, O.G.; Zenin, V.V.; Pugovkina, N.A.; Kozhukharova, I.V.; Borodkina, A.V.; Grinchuk, T.M.; Fridlyanskaya, I.I.; et al. Sublethal heat shock induces premature senescence rather than apoptosis in human mesenchymal stem cells. Cell Stress Chaperones 2014, 19, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Velichko, A.K.; Petrova, N.V.; Razin, S.V.; Kantidze, O.L. Mechanism of heat stress-induced cellular senescence elucidates the exclusive vulnerability of early S-phase cells to mild genotoxic stress. Nucleic Acids Res. 2015, 43, 6309–6320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantidze, O.L.; Velichko, A.K.; Luzhin, A.V.; Razin, S.V. Heat Stress-Induced DNA Damage. Acta Nat. 2016, 8, 75–78. [Google Scholar] [PubMed]
- Vinogradov, A.E.; Shilina, M.A.; Anatskaya, O.V.; Alekseenko, L.L.; Fridlyanskaya, I.I.; Krasnenko, A.; Kim, A.; Korostin, D.; Ilynsky, V.; Elmuratov, A.; et al. Molecular Genetic Analysis of Human Endometrial Mesenchymal Stem Cells That Survived Sublethal Heat Shock. Stem Cells Int. 2017, 2017, 2362630. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.T.; Cesare, A.J.; Rivera, T.; Karlseder, J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015, 522, 492–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erenpreisa, J.; Cragg, M.S. Three steps to the immortality of cancer cells: Senescence, polyploidy and self-renewal. Cancer Cell Int. 2013, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Salmina, K.; Huna, A.; Jackson, T.R.; Vazquez-Martin, A.; Cragg, M.S. The “virgin birth”, polyploidy, and the origin of cancer. Oncoscience 2014, 2, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hopp, N.; Hagen, J.; Aggeler, B.; Kalyuzhny, A.E. Express γ-H2AX Immunocytochemical Detection of DNA Damage. Methods Mol. Biol. 2017, 1644, 123–128. [Google Scholar] [PubMed]
- Hopp, N.; Hagen, J.; Aggeler, B.; Kalyuzhny, A.E. Automated High-Content Screening of γ-H2AX Expression in HeLa Cells. Methods Mol. Biol. 2017, 1554, 273–283. [Google Scholar] [PubMed]
- Roemeling-van Rhijn, M.; de Klein, A.; Douben, H.; Pan, Q.; van der Laan, L.J.; Ijzermans, J.N.; Betjes, M.G.; Baan, C.C.; Weimar, W.; Hoogduijn, M.J. Culture expansion induces non-tumorigenic aneuploidy in adipose tissue-derived mesenchymal stromal cells. Cytotherapy 2013, 15, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Paula, A.C.; Martins, T.M.; Zonari, A.; Frade, S.P.; Angelo, P.C.; Gomes, D.A.; Goes, A.M. Human adipose tissue-derived stem cells cultured in xeno-free culture condition enhance c-MYC expression increasing proliferation but bypassing spontaneous cell transformation. Stem Cell Res. Ther. 2015, 6, 76. [Google Scholar] [CrossRef] [PubMed]
- Zemel’ko, V.I.; Grinchuk, T.M.; Domnina, A.P.; Artsybasheva, I.V.; Zenin, V.V.; Kirsanov, A.A.; Bichevaia, N.K.; Korsak, V.S.; Nikolski, N.N. Multipotent mesenchymal stem cells of desquamated endometrium: Isolation, characterization and use as feeder layer for maintenance of human embryonic stem cell lines. Tsitologiia 2011, 53, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Shilina, M.A.; Grinchuk, T.M.; Nikolsky, N.N. Genetic stability of endometrial mesenchymal stem cells measured by morphological and molecular karyotyping. Tsitologiia 2016, 53, 919–929. [Google Scholar]
- ISCN. ISCN: Intrnation System for Human Cytogenetic Nomenclature; Mitelman, F., Ed.; S. Karger: Basel, Switzerland, 1995; p. 114. [Google Scholar]
- Mamaeva, S.E. Atlas khromosom postoyannykh kletochnykh liny cheloveka i zhivotnykh (Atlas of Chromosomes of Permanent Human and Animal Cell Lines); Nauka: Moscow, Russia, 2002; p. 231. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Somatic polyploidy promotes cell function under stress and energy depletion: Evidence from tissue-specific mammal transcriptome. Funct. Integr. Genom. 2010, 10, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E. Consolidation of slow or fast but not moderately evolving genes at the level of pathways and processes. Gene 2015, 561, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward Nucleic Acids Res. 2015, 43, D1049–D1056.
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E.; Anatskaya, O.V. DNA helix: The importance of being AT-rich. Mamm. Genome 2017, 28, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Chun-Jen Lin, C.; Mo, W.; Dai, H.; Park, Y.Y.; Kim, S.M.; Peng, Y.; Mo, Q.; Siwko, S.; Hu, R.; et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 2014, 5, 3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinogradov, A.E. Bendable genes of warm-blooded vertebrates. Mol. Biol. Evol. 2001, 18, 2195–2200. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Higashino, A.; Takeuchi, K.; Hori, Y.; Koshiba-Takeuchi, K.; Makino, H.; Monobe, Y.; Kishida, M.; Adachi, J.; Takeuchi, J.; et al. Transcriptional dynamics of immortalized human mesenchymal stem cells during transformation. PLoS ONE 2015, 10, e0126562. [Google Scholar]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and safety issues of stem cell-based therapy. Int. J. Med. Sci. 2018, 15, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Rubio, D.; Garcia-Castro, J.; Martin, M.C.; de la Fuente, R.; Cigudosa, J.C.; Lloyd, A.C.; Bernad, A. Spontaneous human adult stem cell transformation. Cancer Res. 2005, 65, 3035–3039. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, A.; Røsland, G.V.; Svendsen, A.; Molven, A.; Immervoll, H.; McCormack, E.; Lønning, P.E.; Primon, M.; Sobala, E.; Tonn, J.C.; et al. Spontaneous malignant transformation of human mesenchymal stem cells reflects cross-contamination: Putting the research field on track-letter. Cancer Res. 2010, 70, 6393–6396. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Mori, E.; Somakos, G.I.; Ohnishi, K.; Ohnishi, T. Heat induces gamma H2AX foci formation in mammalian cells. Mutat. Res. 2008, 656, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Mayshar, Y.; Benvenisty, N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell 2011, 9, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Mayshar, Y.; Benvenisty, N. Significant acquisition of chromosomal aberrations in human adult mesenchymal stem cells: Response to Sensebé et al. Cell Stem Cell 2012, 10, 10–11. [Google Scholar] [CrossRef]
- Borghesi, A.; Avanzini, M.A.; Novara, F.; Mantelli, M.; Lenta, E.; Achille, V.; Cerbo, R.M.; Tzialla, C.; Longo, S.; De Silvestri, A.; et al. Genomic alterations in human umbilical cord derived mesenchymal stromal cells call for stringent quality control before any possible therapeutic approach. Cytotherapy 2013, 15, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.Q.; Zhao, Y.L.; Li, H.C.; Sui, W.G.; Yan, H.C.; Wang, X.Q. Heat stress inhibits proliferation, promotes growth, and induces apoptosis in cultured Lantang swine skeletal muscle satellite cells. J. Zhejiang Univ. Sci. B 2015, 16, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Borgonovo, T.; Vaz, I.M.; Senegaglia, A.C.; Rebelatto, C.L.; Brofman, P.R. Genetic evaluation of mesenchymal stem cells by G-banded karyotyping in a cell technology center. Rev. Bras. Hematol. Hemoter. 2014, 36, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Tarte, K.; Gaillard, J.; Lataillade, J.J.; Fouillard, L.; Becker, M.; Mossafa, H.; Tchirkov, A.; Rouard, H.; Henry, C.; Splingard, M.; et al. Clinical-grade production of human mesenchymal stromal cells: Occurrence of aneuploidy without transformation. Blood 2010, 115, 1549–1553. [Google Scholar] [CrossRef] [PubMed]
- Redaelli, S.; Bentivegna, A.; Foudah, D.; Miloso, M.; Redondo, J.; Riva, G.; Baronchelli, S.; Dalprà, L.; Tredici, G. From cytogenomic to epigenomic profiles: Monitoring the biologic behavior of in vitro cultured human bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 2012, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Enver, T.; Soneji, S.; Joshi, C.; Brown, J.; Iborra, F.; Orntoft, T.; Thykjaer, T.; Maltby, E. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 2005, 14, 3129–3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, D.E.; Harrison, N.J.; Maltby, E.; Smith, K.; Moore, H.D.; Shaw, P.J.; Heath, P.R.; Holden, H.; Andrews, P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007, 25, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015, 16, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardo, M.E.; Zaffaroni, N.; Novara, F.; Cometa, A.M.; Avanzini, M.A.; Moretta, A.; Montagna, D.; Maccario, R.; Villa, R.; Daidone, M.G.; et al. Human bone marrow-derived mesenchymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer 2007, 67, 9142–9149. [Google Scholar]
- Jones, M.; Varella-Garcia, M.; Skokan, M.; Bryce, S.; Schowinsky, J.; Peters, R.; Vang, B.; Brecheisen, M.; Startz, T.; Frank, N.; et al. Genetic stability of bone marrow derived human mesenchymal stromal cells in the Quantum System. Cytotherapy 2013, 15, 1323–1339. [Google Scholar] [CrossRef] [PubMed]
- Roselli, E.A.; Lazzati, S.; Iseppon, F.; Manganini, M.; Marcato, L.; Gariboldi, M.B.; Maggi, F.; Grati, F.R.; Simoni, G. Fetal mesenchymal stromal cells from cryopreserved human chorionic villi: Cytogenetic and molecular analysis of genome stability in long-term cultures. Cytotherapy 2013, 15, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Zaman, W.S.; Makpol, S.; Sathapan, S.; Chua, K.H. Long-term in vitro expansion of human adipose-derived stem cells showed low risk of tumourigenicity. J. Tissue Eng. Regen. Med. 2014, 8, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Velichko, A.K.; Petrova, N.V.; Kantidze, O.L.; Razin, S.V. Dual effect of heat shock on DNA replication and genome integrity. Mol. Biol. Cell 2012, 23, 3450–3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, Y.W.; Wing, M.J.; Xiong, W.; Zhao, J.; Finnell, R.H. Genetic dissection of hyperthermia-induced neural tube defects in mice. Birth Defects Res. A Clin. Mol. Teratol. 2003, 67, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Dreier, J.W.; Andersen, A.M.; Berg-Beckhoff, G. Systematic review and meta-analyses: Fever in pregnancy and health impacts in the offspring. Pediatrics 2014, 133, e674–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anatskaya, O.V.; Vinogradov, A.E. Myocyte ploidy in heart chambers of birds with different locomotor activity. J. Exp. Zool. 2002, 293, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Paradoxical relationship between protein content and nucleolar activity in mammalian cardiomyocytes. Genome 2004, 47, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E.; Kudryavtsev, B.N. Cardiomyocyte ploidy levels in birds with different growth rates. J. Exp. Zool. 2001, 289, 48–58. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Sidorenko, N.V.; Vinogradov, A.E.; Beyer, T.V. Impact of neonatal cryptosporidial gastroenteritis on epigenetic programming of rat hepatocytes. Cell Biol. Int. 2007, 31, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Sidorenko, N.V.; Beyer, T.V.; Vinogradov, A.E. Neonatal cardiomyocyte ploidy reveals critical windows of heart development. Int. J. Cardiol. 2010, 41, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Baryshev, M.; Inashkina, I.; Salmina, K.; Huna, A.; Jackson, T.R.; Erenpreisa, J. DNA methylation of the Oct4A enhancers in embryonal carcinoma cells after etoposide treatment is associated with alternative splicing and altered pluripotency in reversibly senescent cells. Cell Cycle 2018, 17, 362–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez-Martin, A.; Anatskaya, O.V.; Giuliani, A.; Erenpreisa, J.; Huang, S.; Salmina, K.; Inashkina, I.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Somatic polyploidy is associated with the upregulation of c-MYC interacting genes and EMT-like signature. Oncotarget 2016, 7, 75235–75260. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Puzio-Kuter, A.M.; Chan, C.S.; Hainaut, P. The Role of the p53 Protein in Stem-Cell Biology and Epigenetic Regulation. Cold Spring Harb. Respect. Med. 2016, 6, a026153. [Google Scholar] [CrossRef] [PubMed]
- Borodkina, A.V.; Shatrova, A.N.; Deryabin, P.I.; Griukova, A.A.; Abushik, P.A.; Antonov, S.M.; Nikolsky, N.N.; Burova, E.B. Calcium alterations signal either to senescence or to autophagy induction in stem cells upon oxidative stress. Aging (Albany NY) 2016, 8, 3400–3418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanpain, C.; Mohrin, M.; Sotiropoulou, P.A.; Passegué, E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell 2011, 8, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Weeden, C.E.; Asselin-Labat, M.L. Mechanisms of DNA damage repair in adult stem cells and implications for cancer formation. Biochim. Biophys. Acta 2018, 1864, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Buddingh, E.P.; Ruslan, S.E.N.; Reijnders, C.M.A.; Szuhai, K.; Kuijjer, M.L.; Roelofs, H.; Hogendoorn, P.C.W.; Maarten Egeler, R.; Cleton-Jansen, A.M.; Lankester, A.C. Mesenchymal stromal cells of osteosarcoma patients do not show evidence of neoplastic changes during long-term culture. Clin. Sarcoma Res. 2015, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Rigatti, M.J.; Belinsky, G.S.; Godman, C.A.; Giardina, C. DNA damage response to the Mdm2 inhibitor nutlin-3. Biochem. Pharmacol. 2010, 79, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Rigatti, M.J.; Verma, R.; Belinsky, G.S.; Rosenberg, D.W.; Giardina, C. Pharmacological inhibition of Mdm2 triggers growth arrest and promotes DNA breakage in mouse colon tumors and human colon cancer cells. Mol. Carcinog. 2012, 51, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, Y.; Liu, H.; Shi, L.; Sun, X. Icotinib hydrochloride enhances chemo- and radiosensitivity by inhibiting EGFR signaling and attenuating RAD51 expression and function in Hela S3 cells. Onco Targets Ther. 2018, 11, 1245–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husseinzadeh, N.; Husseinzadeh, H.D. mTOR inhibitors and their clinical application in cervical, endometrial and ovarian cancers: A critical review. Gynecol. Oncol. 2014, 133, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Kalimutho, M.; Bain, A.L.; Mukherjee, B.; Nag, P.; Nanayakkara, D.M.; Harten, S.K.; Harris, J.L.; Subramanian, G.N.; Sinha, D.; Shirasawa, S.; et al. Enhanced dependency of KRAS-mutant colorectal cancer cells on RAD51-dependent homologous recombination repair identified from genetic interactions in Saccharomyces cerevisiae. Mol. Oncol. 2017, 11, 470–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, W.; Liu, Q.; Lin, C.C.; Dai, H.; Peng, Y.; Liang, Y.; Peng, G.; Meric-Bernstam, F.; Mills, G.B.; Li, K.; et al. mTOR inhibitors suppress homologous recombination repair and synergize with parp inhibitors via regulating SUV39H1 in BRCA-proficient triple-negative breast cancer. Clin. Cancer Res. 2016, 22, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Gartrell, B.A.; Ying, J.; Sivendran, S.; Boucher, K.M.; Choueiri, T.K.; Sonpavde, G.; Oh, W.K.; Agarwal, N.; Galsky, M.D. Pulmonary complications with the use of mTOR inhibitors in targeted cancer therapy: A systematic review and meta-analysis. Target Oncol. 2014, 9, 195–204. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shilina, M.A.; Grinchuk, T.M.; Anatskaya, O.V.; Vinogradov, A.E.; Alekseenko, L.L.; Elmuratov, A.U.; Nikolsky, N.N. Cytogenetic and Transcriptomic Analysis of Human Endometrial MSC Retaining Proliferative Activity after Sublethal Heat Shock. Cells 2018, 7, 184. https://doi.org/10.3390/cells7110184
Shilina MA, Grinchuk TM, Anatskaya OV, Vinogradov AE, Alekseenko LL, Elmuratov AU, Nikolsky NN. Cytogenetic and Transcriptomic Analysis of Human Endometrial MSC Retaining Proliferative Activity after Sublethal Heat Shock. Cells. 2018; 7(11):184. https://doi.org/10.3390/cells7110184
Chicago/Turabian StyleShilina, Mariia A., Tatiana M. Grinchuk, Olga V. Anatskaya, Alexander E. Vinogradov, Larisa L. Alekseenko, Artem U. Elmuratov, and Nikolai N. Nikolsky. 2018. "Cytogenetic and Transcriptomic Analysis of Human Endometrial MSC Retaining Proliferative Activity after Sublethal Heat Shock" Cells 7, no. 11: 184. https://doi.org/10.3390/cells7110184