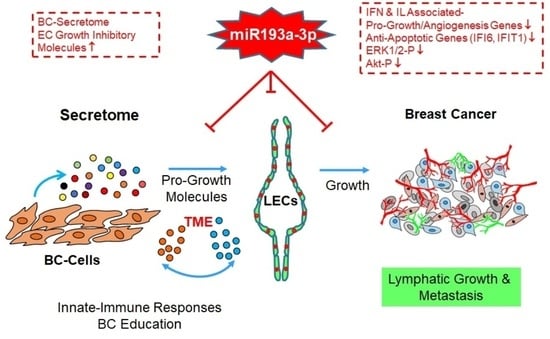
Transcriptomic and Functional Evidence That miRNA193a-3p Inhibits Lymphatic Endothelial Cell (LEC) and LEC + MCF-7 Spheroid Growth Directly and by Altering MCF-7 Secretome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Conditioned Media (CM) Formation
2.3. Spheroid Formation and Culture
2.4. Transfection with miRNA
2.5. Cell Proliferation Assay
2.6. RT-PCR Gene Expression Studies
2.7. Western Blot
2.8. Microarray
2.9. Apoptosis Studies
2.9.1. LEC Apoptosis Studies Using the Proteome Profiler Apoptosis Array
2.9.2. LEC Apoptosis Studies using Hoechst 33342 and PI Staining
2.10. Cytokine Proteome Array in the Secretome
2.11. Statistical Analysis
3. Results
3.1. miR193a-3p Inhibits the Growth in LECs and LECs-MCF-7 Spheroids
3.2. Modulatory Effects of miRNA193a-3p on ERK-MAPK/PI3K-Akt Phosphorylation in Spheroids
3.3. The Effect of Conditioned Media from miR193a-3p Transfected MCF-7 cells on LEC Growth and The Role of ERK-MAPK/PI3K-Akt Pathways
3.4. Estrogen Promotes LEC Growth by Downregulating miRNA193a-3p Expression
3.5. Effects of E2 on LEC + MCF-7 Spheroid Growth and Activation of ERK-MAPK/PI3K-Akt Pathways: Modulatory Role of miR193a-3p
3.6. Microarray Analysis
3.6.1. Differentially Regulated Genes
3.6.2. Pathway Enrichment Analysis of DRGs
3.6.3. Gene Identification in Biological Pathways
3.6.4. Genes Involved in Apoptotic Pathways
3.6.5. miRNA193a-3p Induces LEC Apoptosis
3.7. Cytokine Proteome Profiling of MCF-7 Secretome
3.8. Secretome from miR193a-3p Transfected MCF-7 Cells Induces Death/Apoptosis in LECs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Libson, S.; Lippman, M. A review of clinical aspects of breast cancer. Int. Rev. Psychiatry 2014, 26, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, L.; Jordāo, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor Microenvironment in Breast Cancer-Updates on Therapeutic Implications and Pathologic Assessment. Cancers 2021, 13, 4233. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Pandey, N.B.; Popel, A.S. Crosstalk between cancer cells and blood endothelial and lymphatic endothelial cells in tumour and organ microenvironment. Expert Rev. Mol. Med. 2015, 17, e3. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madu, C.O.; Wang, S.; Madu, C.O.; Lu, Y. Angiogenesis in Breast Cancer Progression, Diagnosis, and Treatment. J. Cancer 2020, 11, 4474–4494. [Google Scholar] [CrossRef]
- Jones, D. Parallels of Resistance between Angiogenesis and Lymphangiogenesis Inhibition in Cancer Therapy. Cells 2020, 9, 762. [Google Scholar] [CrossRef] [Green Version]
- Ayoub, N.M.; Jaradat, S.K.; Al-Shami, K.M.; Alkhalifa, A.E. Targeting Angiogenesis in Breast Cancer: Current Evidence and Future Perspectives of Novel Anti-Angiogenic Approaches. Front. Pharmacol. 2022, 13, 838133. [Google Scholar] [CrossRef]
- Aalders, K.C.; Tryfonidis, K.; Senkus, E.; Cardoso, F. Anti-angiogenic treatment in breast cancer: Facts, successes, failures and future perspectives. Cancer Treat. Rev. 2017, 53, 98–110. [Google Scholar] [CrossRef]
- Ansari, M.J.; Bokov, D.; Markov, A.; Jalil, A.T.; Shalaby, M.N.; Suksatan, W.; Chupradit, S.; Al-Ghamdi, H.S.; Shomali, N.; Zamani, A.; et al. Cancer combination therapies by angiogenesis inhibitors; A comprehensive review. Cell Commun. Signal. 2022, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Kayani, M.; Kayani, M.A.; Malik, F.A.; Faryal, R. Role of miRNAs in breast cancer. Asian Pac. J. Cancer Prev. 2011, 12, 3175–3180. [Google Scholar] [PubMed]
- Fridrichova, I.; Zmetakova, I. MicroRNAs Contribute to Breast Cancer Invasiveness. Cells 2019, 8, 1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Khordadmehr, M.; Shahbazi, R.; Ezzati, H.; Jigari-Asl, F.; Sadreddini, S.; Baradaran, B. Key microRNAs in the biology of breast cancer; emerging evidence in the last decade. J. Cell Physiol. 2019, 234, 8316–8326. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; de Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. eJIFCC 2019, 30, 114–127. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Tsai, K.W.; Leung, C.M.; Lo, Y.H.; Chen, T.W.; Chan, W.C.; Yu, S.Y.; Tu, Y.T.; Lam, H.C.; Li, S.C.; Ger, L.P.; et al. Arm Selection Preference of MicroRNA-193a Varies in Breast Cancer. Sci. Rep. 2016, 6, 28176. [Google Scholar] [CrossRef] [PubMed]
- Khordadmehr, M.; Shahbazi, R.; Baradaran, B.; Sadreddini, S.; Shanebandi, D.; Hajiasgharzadeh, K. Restoring of miR-193a-5p Sensitizes Breast Cancer Cells to Paclitaxel through P53 Pathway. Adv. Pharm. Bull. 2020, 10, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yan, M.; Yu, T.; Ge, H.; Lin, H.; Li, J.; Liu, Y.; Geng, Q.; Zhu, M.; Liu, L.; et al. Quantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancer. Cell Physiol. Biochem. 2015, 35, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Takahashi, M.; Ohnuma, S.; Unno, M.; Yoshino, Y.; Ouchi, K.; Takahashi, S.; Yamada, Y.; Shimodaira, H.; Ishioka, C. microRNA-193a-3p is specifically down-regulated and acts as a tumor suppressor in BRAF-mutated colorectal cancer. BMC Cancer 2017, 17, 723. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.N.; Lin, J.; Li, Y.H.; Gao, L.; Wang, X.R.; Wang, W.; Kang, H.Y.; Yan, G.T.; Wang, L.L.; Yu, L. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene 2011, 30, 3416–3428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossi, I.; Salvi, A.; Abeni, E.; Marchina, E.; de Petro, G. Biological Function of MicroRNA193a-3p in Health and Disease. Int. J. Genom. 2017, 2017, 5913195. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, H. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J. Biochem. Mol. Toxicol. 2018, 32, e22008. [Google Scholar] [CrossRef]
- Van den Bosch, M.T.J.; Yahyanejad, S.; Alemdehy, M.F.; Telford, B.J.; de Gunst, T.; den Boer, H.C.; Vos, R.M.; Stegink, M.; van Pinxteren, L.A.H.; Schaapveld, R.Q.J.; et al. Transcriptome-wide analysis reveals insight into tumor suppressor functions of 1B3, a novel synthetic miR-193a-3p mimic. Mol. Ther. Nucleic Acids 2021, 23, 1161–1171. [Google Scholar] [CrossRef]
- Azzarito, G.; Kurmann, L.; Leeners, B.; Dubey, R.K. Micro-RNA193a-3p Inhibits Breast Cancer Cell Driven Growth of Vascular Endothelial Cells by Altering Secretome and Inhibiting Mitogenesis: Transcriptomic and Functional Evidence. Cells 2022, 11, 2967. [Google Scholar] [CrossRef]
- Lee, E.; Fertig, E.J.; Jin, K.; Sukumar, S.; Pandey, N.B.; Popel, A.S. Breast cancer cells condition lymphatic endothelial cells within pre-metastatic niches to promote metastasis. Nat. Commun. 2014, 5, 4715. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.M.; Luo, B.; Ma, R.; Luo, X.X.; Chen, Y.S.; Li, Y. Lymphatic Endothelial Markers and Tumor Lymphangiogenesis Assessment in Human Breast Cancer. Diagnostics 2021, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Veikkola, T.; Lohela, M.; Ikenberg, K.; Mäkinen, T.; Korff, T.; Saaristo, A.; Petrova, T.; Jeltsch, M.; Augustin, H.G.; Alitalo, K. Intrinsic versus microenvironmental regulation of lymphatic endothelial cell phenotype and function. FASEB J. 2003, 17, 2006–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travis, R.C.; Key, T.J. Oestrogen exposure and breast cancer risk. Breast Cancer Res. 2003, 5, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Klinge, C.M. miRNAs regulated by estrogens, tamoxifen, and endocrine disruptors and their downstream gene targets. Mol. Cell Endocrinol. 2015, 418 Pt 3, 273–297. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Xu, J.; Qi, J.; Zhang, L.; Wang, J.; Liang, J.; Qian, N.; Zhou, H.; Wei, L.; Deng, L. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J. Cell Sci. 2013, 126 Pt 4, 978–988. [Google Scholar] [CrossRef] [Green Version]
- Lv, H.; Sun, Y.; Zhang, Y. MiR-133 is Involved in Estrogen Deficiency-Induced Osteoporosis through Modulating Osteogenic Differentiation of Mesenchymal Stem Cells. Med. Sci. Monit. 2015, 21, 1527–1534. [Google Scholar]
- Fu, J.; Hao, L.; Tian, Y.; Liu, Y.; Gu, Y.; Wu, J. miR-199a-3p is involved in estrogen-mediated autophagy through the IGF-1/mTOR pathway in osteocyte-like MLO-Y4 cells. J. Cell Physiol. 2018, 233, 2292–2303. [Google Scholar] [CrossRef] [PubMed]
- Vrtačnik, P.; Ostanek, B.; Mencej-Bedrač, S.; Marc, J. The many faces of estrogen signaling. Biochem. Med. 2014, 24, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef] [Green Version]
- Wickramasinghe, N.S.; Manavalan, T.T.; Dougherty, S.M.; Riggs, K.A.; Li, Y.; Klinge, C.M. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009, 37, 2584–2595. [Google Scholar] [CrossRef]
- Evangelista, A.F.; Oliveira, R.J.; Silva, V.A.O.; Vieira, R.A.D.C.; Reis, R.M.; Marques, M.M.C. Integrated analysis of mRNA and miRNA profiles revealed the role of miR-193 and miR-210 as potential regulatory biomarkers in different molecular subtypes of breast cancer. BMC Cancer 2021, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, S.; Wang, M.; Liu, D.; Liu, Y.; Zhang, Y.; Zhang, Q. Epigenetically altered miR-193a-3p promotes HER2 positive breast cancer aggressiveness by targeting GRB7. Int. J. Mol. Med. 2019, 43, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzarito, G.; Szutkowska, M.E.; Saltari, A.; Jackson, E.K.; Leeners, B.; Rosselli, M.; Dubey, R.K. Mammary Epithelial and Endothelial Cell Spheroids as a Potential Functional In vitro Model for Breast Cancer Research. J. Vis. Exp. 2021, 173, e62940. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Khordadmehr, M.; Shahbazi, R.; Sadreddini, S.; Baradaran, B. miR-193: A new weapon against cancer. J. Cell Physiol. 2019, 234, 16861–16872. [Google Scholar] [CrossRef]
- Sheng, S.; Qiao, M.; Pardee, A.B. Metastasis and AKT activation. J. Cell Physiol. 2009, 218, 451–454. [Google Scholar] [CrossRef]
- Li, Q.T.; Feng, Y.M.; Ke, Z.H.; Qiu, M.J.; He, X.X.; Wang, M.M.; Li, Y.N.; Xu, J.; Shi, L.L.; Xiong, Z.F. KCNN4 promotes invasion and metastasis through the MAPK/ERK pathway in hepatocellular carcinoma. J. Investig. Med. 2020, 68, 68–74. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.Y.; Fattet, L.; Yang, J. Molecular pathways: Linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin. Cancer Res. 2015, 21, 962–968. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Pandey, N.B.; Popel, A.S. Lymphatic endothelial cells support tumor growth in breast cancer. Sci. Rep. 2014, 4, 5853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzarito, G.; Visentin, M.; Leeners, B.; Dubey, R.K. Transcriptomic and Functional Evidence for Differential Effects of MCF-7 Breast Cancer Cell-Secretome on Vascular and Lymphatic Endothelial Cell Growth. Int. J. Mol. Sci. 2022, 23, 7192. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, H.; Liu, Z.Y.; Zhu, Q.; Shan, C.; Zhang, K.Q. Advanced glycation end products induce the apoptosis of and inflammation in mouse podocytes through CXCL9-mediated JAK2/STAT3 pathway activation. Int. J. Mol. Med. 2017, 40, 1185–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Yang, X.G.; Wang, F.; Ma, X.Y. IL-1α induces apoptosis and inhibits the osteoblast differentiation of MC3T3-E1 cells through the JNK and p38 MAPK pathways. Int. J. Mol. Med. 2016, 38, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Hao, J.; Shi, Y. IL23 suppresses proliferation and promotes apoptosis of human granulosa-like tumor cell line KGN by targeting the androgen receptor signal pathway. Gynecol. Endocrinol. 2022, 38, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Meng, J.; Zhang, C.; Duan, Y.; Zhao, L.; Wang, S.; Shan, B. Effects on apoptosis and cell cycle arrest contribute to the antitumor responses of interleukin-27 mediated by retrovirus in human pancreatic carcinoma cells. Oncol. Rep. 2012, 27, 1497–1503. [Google Scholar] [PubMed] [Green Version]
- Persaud, L.; Mighty, J.; Zhong, X.; Francis, A.; Mendez, M.; Muharam, H.; Redenti, S.M.; Das, D.; Aktas, B.H.; Sauane, M. IL-24 Promotes Apoptosis through cAMP-Dependent PKA Pathways in Human Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3561. [Google Scholar] [CrossRef] [Green Version]
- Zi, M.; Xu, Y. Involvement of cystatin C in immunity and apoptosis. Immunol. Lett. 2018, 196, 80–90. [Google Scholar] [CrossRef]
- Nabata, A.; Kuroki, M.; Ueba, H.; Hashimoto, S.; Umemoto, T.; Wada, H.; Yasu, T.; Saito, M.; Momomura, S.I.; Kawakami, M. C-reactive protein induces endothelial cell apoptosis and matrix metalloproteinase-9 production in human mononuclear cells: Implications for the destabilization of atherosclerotic plaque. Atherosclerosis 2008, 196, 129–135. [Google Scholar] [CrossRef]
- Kasibhatla, S.; Jessen, K.A.; Maliartchouk, S.; Wang, J.Y.; English, N.M.; Drewe, J.; Qiu, L.; Archer, S.P.; Ponce, A.E.; Sirisoma, N.; et al. A role for transferrin receptor in triggering apoptosis when targeted with gambogic acid. Proc. Natl. Acad. Sci. USA 2005, 102, 12095–12100. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Strawn, T.L.; Luo, M.; Wang, L.; Li, R.; Ren, M.; Xia, J.; Zhang, Z.; Ma, W.; Luo, T.; et al. Plasminogen activator inhibitor-1 inhibits angiogenic signaling by uncoupling vascular endothelial growth factor receptor-2-αVβ3 integrin cross talk. Arterioscler Thromb. Vasc. Biol. 2015, 35, 111–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakazato, T.; Sagawa, M.; Yamato, K.; Xian, M.; Yamamoto, T.; Suematsu, M.; Ikeda, Y.; Kizaki, M. Myeloperoxidase is a key regulator of oxidative stress mediated apoptosis in myeloid leukemic cells. Clin. Cancer Res. 2007, 13 Pt 1, 5436–5445. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020, 20, 173–185. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Cao, Z.; Liao, Q.; Su, M.; Huang, K.; Jin, J.; Cao, D. AKT and ERK dual inhibitors: The way forward? Cancer Lett. 2019, 459, 30–40. [Google Scholar] [CrossRef]
- Saleiro, D.; Platanias, L.C. Interferon signaling in cancer. Non-canonical pathways and control of intracellular immune checkpoints. Semin. Immunol. 2019, 43, 101299. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Peng, L.; Zhang, Y.; Liu, Z.; Li, W.; Chen, S.; Li, G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 2017, 34, 101. [Google Scholar] [CrossRef] [Green Version]
- Forey, P.; Cros-Perrial, E.; Dumontet, C.; Jordheim, L.P. Targeting the Nucleotide Metabolism Proteins of the NUDIX Family and SAMHD1 in Cancer. Curr. Med. Chem. 2021, 28, 4088–4116. [Google Scholar] [CrossRef]
- Huang, P.; Liao, R.; Chen, X.; Wu, X.; Li, X.; Wang, Y.; Cao, Q.; Dong, C. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics 2020, 10, 4644–4658. [Google Scholar] [CrossRef]
- Pidugu, V.K.; Pidugu, H.B.; Wu, M.M.; Liu, C.J.; Lee, T.C. Emerging Functions of Human IFIT Proteins in Cancer. Front. Mol. Biosci. 2019, 6, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henle, A.M.; Nassar, A.; Puglisi-Knutson, D.; Youssef, B.; Knutson, K.L. Downregulation of TAP1 and TAP2 in early stage breast cancer. PLoS ONE 2017, 12, e0187323. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.; Cheng, Y.C.; Tsai, W.C.; Chen, Y. PSMB8 inhibition decreases tumor angiogenesis in glioblastoma through vascular endothelial growth factor A reduction. Cancer Sci. 2020, 111, 4142–4153. [Google Scholar] [CrossRef] [PubMed]
- Rouette, A.; Trofimov, A.; Haberl, D.; Boucher, G.; Lavallée, V.P.; D’Angelo, G.; Hébert, J.; Sauvageau, G.; Lemieux, S.; Perreault, C. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci. Rep. 2016, 6, 34019. [Google Scholar] [CrossRef] [Green Version]
- Cheriyath, V.; Kaur, J.; Davenport, A.; Khalel, A.; Chowdhury, N.; Gaddipati, L. G1P3 (IFI6), a mitochondrial localised antiapoptotic protein, promotes metastatic potential of breast cancer cells through mtROS. Br. J. Cancer 2018, 119, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Qiu, X.; Lin, S.; Chen, X.; Wang, T.; Liao, T. Knockdown of IFI27 inhibits cell proliferation and invasion in oral squamous cell carcinoma. World J. Surg. Oncol. 2018, 16, 64. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, C. Prognostic characterization of OAS1/OAS2/OAS3/OASL in breast cancer. BMC Cancer 2020, 20, 575. [Google Scholar] [CrossRef]
- Friedlová, N.; Zavadil Kokáš, F.; Hupp, T.R.; Vojtěšek, B.; Nekulová, M. IFITM protein regulation and functions: Far beyond the fight against viruses. Front. Immunol. 2022, 13, 1042368. [Google Scholar] [CrossRef]
- Popson, S.A.; Hughes, C.C.W. A role for IFITM proteins in angiogenesis. FASEB J. 2010, 24, 750–751. [Google Scholar] [CrossRef]
- Li, L.; Bai, J.; Fan, H.; Yan, J.; Li, S.; Jiang, P. E2 ubiquitin-conjugating enzyme UBE2L6 promotes Senecavirus A proliferation by stabilizing the viral RNA polymerase. PLoS Pathog. 2020, 16, e1008970. [Google Scholar] [CrossRef]
- Tan, Y.; Zhou, G.; Wang, X.; Chen, W.; Gao, H. USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int. J. Oncol. 2018, 53, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29. [Google Scholar] [PubMed]
- Tang, J.; Yang, Q.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Weighted gene correlation network analysis identifies RSAD2, HERC5, and CCL8 as prognostic candidates for breast cancer. J. Cell Physiol. 2020, 235, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Li, Y.; Zhang, Y.; Zhang, L.; Wang, Z.; Zhang, X.; Gui, L.; Huang, J. IFI6 Inhibits Apoptosis via Mitochondrial-Dependent Pathway in Dengue Virus 2 Infected Vascular Endothelial Cells. PLoS ONE 2015, 10, e0132743. [Google Scholar]
- Zhang, X.; Valenzuela, N.M.; Reed, E.F. HLA class I antibody-mediated endothelial and smooth muscle cell activation. Curr Opin. Organ Transplant. 2012, 17, 446–451. [Google Scholar] [CrossRef] [Green Version]
- Ghavami, S.; Eshragi, M.; Ande, S.R.; Chazin, W.J.; Klonisch, T.; Halayko, A.J.; McNeill, K.D.; Hashemi, M.; Kerkhoff, C.; Los, M. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010, 20, 314–331. [Google Scholar] [CrossRef] [Green Version]
- Augustin, S.; Berard, M.; Kellaf, S.; Peyri, N.; Fauvel-Lafève, F.; Legrand, C.; He, L.; Crépin, M. Matrix metalloproteinases are involved in both type I (apoptosis) and type II (autophagy) cell death induced by sodium phenylacetate in MDA-MB-231 breast tumour cells. Anticancer Res. 2009, 29, 1335–1343. [Google Scholar]
- Yang, S.D.; Yang, D.L.; Sun, Y.P.; Wang, B.L.; Ma, L.; Feng, S.Q.; Ding, W.Y. 17β-estradiol protects against apoptosis induced by interleukin-1β in rat nucleus pulposus cells by down-regulating MMP-3 and MMP-13. Apoptosis 2015, 20, 348–357. [Google Scholar] [CrossRef]
- Taira, N.; Yamaguchi, T.; Kimura, J.; Lu, Z.G.; Fukuda, S.; Higashiyama, S.; Ono, M.; Yoshida, K. Induction of amphiregulin by p53 promotes apoptosis via control of microRNA biogenesis in response to DNA damage. Proc. Natl. Acad. Sci. USA 2014, 111, 717–722. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Lee, E.; Sun, Z.; Wang, X.; Ren, T.; Zou, Z.; Jin, J.; Li, J.; Zhang, J.; Li, Y.; et al. Perilipin 2 Impacts Acute Kidney Injury via Regulation of PPARα. J. Immunol. Res. 2021, 2021, 9972704. [Google Scholar] [CrossRef]
- Yun, H.; Xie, J.; Olumi, A.F.; Ghosh, R.; Kumar, A.P. Activation of AKR1C1/ERβ induces apoptosis by downregulation of c-FLIP in prostate cancer cells: A prospective therapeutic opportunity. Oncotarget 2015, 6, 11600–11613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pidugu, V.K.; Wu, M.M.; Yen, A.H.; Pidugu, H.B.; Chang, K.W.; Liu, C.J.; Lee, T.C. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 2019, 38, 3232–3247. [Google Scholar] [CrossRef] [PubMed]
- Ogony, J.; Choi, H.J.; Lui, A.; Cristofanilli, M.; Lewis-Wambi, J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Walter, K.R.; Goodman, M.L.; Singhal, H.; Hall, J.A.; Li, T.; Holloran, S.M.; Trinca, G.M.; Gibson, K.A.; Jin, V.X.; Greene, G.L.; et al. Interferon-Stimulated Genes Are Transcriptionally Repressed by PR in Breast Cancer. Mol. Cancer Res. 2017, 15, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Popson, S.A.; Ziegler, M.E.; Chen, X.; Holderfield, M.T.; Shaaban, C.I.; Fong, A.H.; Welch-Reardon, K.M.; Papkoff, J.; Hughes, C.C. Interferon-induced transmembrane protein 1 regulates endothelial lumen formation during angiogenesis. Arterioscler. Thromb Vasc. Biol. 2014, 34, 1011–1019. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.S.; Zhao, X.; Cheng, X.; Guan, D.; Mahabeleshwar, G.H.; Liu, Y.; Borden, E.; Jain, M.K.; Kao, H.Y. Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon α-mediated inhibition of angiogenesis. J. Biol. Chem. 2017, 292, 10048–10060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccarese, F.; Grassi, A.; Pasqualini, L.; Rosano, S.; Noghero, A.; Montenegro, F.; Bussolino, F.; Di Camillo, B.; Finesso, L.; Toffolo, G.M.; et al. Genetic perturbation of IFN-α transcriptional modulators in human endothelial cells uncovers pivotal regulators of angiogenesis. Comput. Struct. Biotechnol. J. 2020, 18, 3977–3986. [Google Scholar] [CrossRef]
- Burkart, C.; Arimoto, K.; Tang, T.; Cong, X.; Xiao, N.; Liu, Y.C.; Kotenko, S.V.; Ellies, L.G.; Zhang, D.E. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 2013, 5, 1035–1050. [Google Scholar] [CrossRef]
- Cheon, H.; Wang, Y.; Wightman, S.M.; Jackson, M.W.; Stark, G.R. How cancer cells make and respond to interferon-I. Trends Cancer 2022, 9, 83–92. [Google Scholar] [CrossRef]
- Chiang, K.C.; Huang, S.T.; Wu, R.C.; Huang, S.C.; Yeh, T.S.; Chen, M.H.; Hsu, J.T.; Chen, L.W.; Kuo, S.F.; Chueh, H.Y.; et al. Interferon α-inducible protein 27 is an oncogene and highly expressed in cholangiocarcinoma patients with poor survival. Cancer Manag. Res. 2019, 11, 1893–1905. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zu, T.; Li, T.; Li, M.; Mi, J.; Bai, F.; Liu, G.; Wen, J.; Li, H.; Brakebusch, C.; et al. ATF3 downmodulates its new targets IFI6 and IFI27 to suppress the growth and migration of tongue squamous cell carcinoma cells. PLoS Genet. 2021, 17, e1009283. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhang, H.; Long, Y.; Hua, H.; Jiang, Y.; Jing, J. PARP9 is overexpressed in human breast cancer and promotes cancer cell migration. Oncol. Lett. 2018, 16, 4073–4077. [Google Scholar] [CrossRef] [Green Version]
- Kolben, T.; Jeschke, U.; Reimer, T.; Karsten, N.; Schmoeckel, E.; Semmlinger, A.; Mahner, S.; Harbeck, N.; Kolben, T.M. Induction of apoptosis in breast cancer cells in vitro by Fas ligand reverse signaling. J. Cancer Res. Clin. Oncol. 2018, 144, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Riggins, R.B.; Mulla, J.E.; Chung, H.; Zwart, A.; Clarke, R. IFNgamma restores breast cancer sensitivity to fulvestrant by regulating STAT1, IFN regulatory factor 1, NF-kappaB, BCL2 family members, and signaling to caspase-dependent apoptosis. Mol. Cancer Ther. 2010, 9, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Langer, H.F.; Chung, K.J.; Orlova, V.V.; Choi, E.Y.; Kaul, S.; Kruhlak, M.J.; Alatsatianos, M.; DeAngelis, R.A.; Roche, P.A.; Magotti, P.; et al. Complement-mediated inhibition of neovascularization reveals a point of convergence between innate immunity and angiogenesis. Blood 2010, 116, 4395–4403. [Google Scholar] [CrossRef] [Green Version]
- Goda, C.; Kanaji, T.; Kanaji, S.; Tanaka, G.; Arima, K.; Ohno, S.; Izuhara, K. Involvement of IL-32 in activation-induced cell death in T cells. Int. Immunol. 2006, 18, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Bossila, E.A.; Ma, X.; Zhao, C.; Zhao, Y. Dual Immune Regulatory Roles of Interleukin-33 in Pathological Conditions. Cells 2022, 11, 3237. [Google Scholar] [CrossRef]
- Theodoropoulou, S.; Copland, D.A.; Liu, J.; Wu, J.; Gardner, P.J.; Ozaki, E.; Doyle, S.L.; Campbell, M.; Dick, A.D. Interleukin-33 regulates tissue remodelling and inhibits angiogenesis in the eye. J. Pathol. 2017, 241, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Ma, J.; Su, H.; Guo, T.; Cai, H.; Chen, Q.; Zhao, X.; Qi, J.; Du, J. Interleukin-1 receptor antagonist inhibits angiogenesis in gastric cancer. Int. J. Clin. Oncol. 2018, 23, 659–670. [Google Scholar] [CrossRef] [Green Version]
- Lasagni, L.; Francalanci, M.; Annunziato, F.; Lazzeri, E.; Giannini, S.; Cosmi, L.; Sagrinati, C.; Mazzinghi, B.; Orlando, C.; Maggi, E.; et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 2003, 197, 1537–1549. [Google Scholar] [CrossRef]












| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| MMP1 | matrix metallopeptidase 1 | 3.06 | 6.61 × 10−8 |
| S100A7 | S100 calcium-binding protein A7 | 2.73 | 3.08 × 10−7 |
| S100A8 | S100 calcium-binding protein A8 | 2.26 | 2.83 × 10−6 |
| CEMIP | cell migration-inducing protein, hyaluronan binding | 2.01 | 1.25 × 10−6 |
| MMP13 | matrix metallopeptidase 13 | 1.93 | 5.61 × 10−6 |
| AKR1C1 | aldo-keto reductase family 1, member C1 | 1.7 | 8.07 × 10−5 |
| CA9 | carbonic anhydrase IX | 1.63 | 8.52 × 10−5 |
| AREG | amphiregulin | 1.61 | 0.0065 |
| HMOX1 | heme oxygenase 1 | 1.55 | 0.0016 |
| PLIN2 | perilipin 2 | 1.53 | 0.01 |
| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| IFI6 | interferon, alpha-inducible protein 6 | −4.84 | 9.24 × 10−8 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | −4.29 | 5.98 × 10−8 |
| OAS1 | 2-5-oligoadenylate synthetase 1 | −3.20 | 2.65 × 10−6 |
| PSMB9 | proteasome subunit beta 9 | −2.95 | 8.75 × 10−7 |
| HLA-B | major histocompatibility complex, class I, B | −2.78 | 9.24 × 10−8 |
| OAS2 | 2-5-oligoadenylate synthetase 2 | −2.53 | 2.65 × 10−6 |
| HLA-A | major histocompatibility complex, class I, A | −2.51 | 3.08 × 10−7 |
| IFITM1 | interferon induced transmembrane protein 1 | −2.44 | 1.58 × 10−7 |
| PARP9 | poly(ADP-ribose) polymerase family member 9 | −2.42 | 7.67 × 10−6 |
| PSMB8 | proteasome subunit beta 8 | −2.41 | 2.52 × 10−6 |
| Pathway | Overlap | Adj. p-Value |
|---|---|---|
| BioPlanet | ||
| Interferon alpha/beta signaling | 20/64 | 1.22 × 10−26 |
| Interferon signaling | 26/168 | 1.22 × 10−26 |
| Immune system signaling by interferons, interleukins, prolactin, and growth hormones | 26/280 | 6.36 × 10−21 |
| Immune system | 35/998 | 4.68 × 10−15 |
| Interferon gamma signaling pathway | 13/97 | 3.76 × 10−12 |
| Type II interferon signaling (interferon gamma) | 9/50 | 1.37 × 10−9 |
| Antiviral mechanism by interferon-stimulated genes | 7/70 | 8.22 × 10−6 |
| Interleukin-1 regulation of extracellular matrix | 8/120 | 2.13 × 10−5 |
| Adaptive immune system | 15/606 | 1.29 × 10−4 |
| Interleukin-2 signaling pathway | 17/847 | 4.18 × 10−4 |
| ABC transporters | 4/47 | 0.0032 |
| Interleukin-12-mediated signaling events | 04/65 | 0.0094 |
| Prolactin regulation of apoptosis | 5/118 | 0.0102 |
| Integrated cancer pathway | 3/35 | 0.0127 |
| Interleukin-4 regulation of apoptosis | 7/267 | 0.0142 |
| Interferon alpha signaling pathway | 2/10 | 0.0145 |
| Integrated breast cancer pathway | 5/152 | 0.0230 |
| Interleukin-5 signaling pathway | 3/49 | 0.0278 |
| Innate immune system | 7/319 | 0.0316 |
| Interleukin-4 signaling pathway | 4/104 | 0.0316 |
| Interleukin-10 anti-inflammatory signaling pathway | 2/16 | 0.0322 |
| Apoptotic signaling in response to DNA damage | 2/20 | 0.0456 |
| Estrogen receptor signaling pathway | 2/20 | 0.0457 |
| GO Biological Process | ||
| Cellular response to type I interferon (GO:0071357) | 22/65 | 4.66 × 10−30 |
| Type I interferon signaling pathway (GO:0060337) | 22/65 | 4.66 × 10−30 |
| Cytokine-mediated signaling pathway (GO:0019221) | 30/621 | 4.83 × 10−16 |
| Interferon gamma-mediated signaling pathway (GO:0060333) | 13/68 | 5.37 × 10−14 |
| Cellular response to interferon gamma (GO:0071346) | 14/121 | 5.05 × 10−12 |
| Regulation of immune response (GO:0050776) | 13/179 | 1.06 × 10−8 |
| Response to interferon beta (GO:0035456) | 5/28 | 4.74 × 10−5 |
| Negative regulation of cytokine-mediated signaling pathway (GO:0001960) | 6/55 | 6.89 × 10−5 |
| Positive regulation of cytokine production involved in immune response (GO:0002720) | 5/33 | 9.83 × 10−5 |
| Interleukin-27-mediated signaling pathway (GO:0070106) | 4/15 | 9.83 × 10−5 |
| Negative regulation of type I interferon-mediated signaling pathway (GO:0060339) | 4/16 | 1.18 × 10−4 |
| Positive regulation of interferon beta production (GO:0032728) | 5/36 | 1.32 × 10−4 |
| Innate immune response (GO:0045087) | 11/302 | 1.53 × 10-4 |
| Positive regulation of type I interferon production (GO:0032481) | 6/77 | 3.67 × 10−4 |
| Regulation of interferon beta production (GO:0032648) | 5/49 | 5.17 × 10−4 |
| Positive regulation of chemokine production (GO:0032722) | 5/53 | 7.26 × 10−4 |
| Positive regulation of apoptotic signaling pathway (GO:2001235) | 5/59 | 0.0011 |
| Regulation of type I interferon-mediated signaling pathway (GO:0060338) | 4/30 | 0.0011 |
| Regulation of innate immune response (GO:0045088) | 5/64 | 0.0015 |
| Regulation of chemokine (C-X-C motif) ligand 2 production (GO:2000341) | 3/13 | 0.0019 |
| Antiviral innate immune response (GO:0140374) | 3/14 | 0.0023 |
| Negative regulation of innate immune response (GO:0045824) | 4/38 | 0.0025 |
| Positive regulation of immune response (GO:0050778) | 5/75 | 0.0028 |
| Positive regulation of tumor necrosis factor production (GO:0032760) | 5/77 | 0.0031 |
| Regulation of tumor necrosis factor production (GO:0032680) | 6/124 | 0.0033 |
| Negative regulation of extrinsic apoptotic signaling pathway (GO:2001237) | 5/80 | 0.0035 |
| Negative regulation of type I interferon production (GO:0032480) | 4/43 | 0.0035 |
| Negative regulation of chemokine production (GO:0032682) | 3/17 | 0.0035 |
| Positive regulation of tumor necrosis factor superfamily cytokine production (GO:1903557) | 5/81 | 0.0036 |
| Response to interferon alpha (GO:0035455) | 3/18 | 0.0039 |
| Positive regulation of cell population proliferation (GO:0008284) | 11/474 | 0.0044 |
| Positive regulation of interferon alpha production (GO:0032727) | 3/20 | 0.0050 |
| Regulation of response to cytokine stimulus (GO:0060759) | 3/22 | 0.0065 |
| Response to cytokine (GO:0034097) | 6/150 | 0.0071 |
| Regulation of interferon gamma-mediated signaling pathway (GO:0060334) | 3/23 | 0.0072 |
| Regulation of interferon alpha production (GO:0032647) | 3/25 | 0.0090 |
| Apoptotic process (GO:0006915) | 7/231 | 0.0114 |
| Negative regulation of cytokine production (GO:0001818) | 6/182 | 0.0151 |
| Apoptotic mitochondrial changes (GO:0008637) | 3/33 | 0.0161 |
| Negative regulation of extrinsic apoptotic signaling pathway via death domain receptors (GO:1902042) | 3/34 | 0.0174 |
| Positive regulation of cytokine production (GO:0001819) | 8/335 | 0.0181 |
| Cellular response to tumor necrosis factor (GO:0071356) | 6/194 | 0.0192 |
| Response to interferon gamma (GO:0034341) | 4/80 | 0.0210 |
| Positive regulation of intrinsic apoptotic signaling pathway (GO:2001244) | 3/40 | 0.0237 |
| Regulation of chemokine production (GO:0032642) | 3/42 | 0.0262 |
| Regulation of extrinsic apoptotic signaling pathway via death domain receptors (GO:1902041) | 3/42 | 0.0262 |
| Regulation of type I interferon production (GO:0032479) | 4/89 | 0.0269 |
| Regulation of cytokine production (GO:0001817) | 5/150 | 0.0283 |
| Regulation of endothelial cell apoptotic process (GO:2000351) | 3/44 | 0.0284 |
| Regulation of response to interferon gamma (GO:0060330) | 2/14 | 0.0330 |
| Positive regulation of defense response (GO:0031349) | 4/98 | 0.0331 |
| Cellular response to cytokine stimulus (GO:0071345) | 9/482 | 0.0341 |
| Positive regulation of response to cytokine stimulus (GO:0060760) | 2/15 | 0.0356 |
| Regulation of ERK1 and ERK2 cascade (GO:0070372) | 6/238 | 0.0368 |
| Regulation of intrinsic apoptotic signaling pathway (GO:2001242) | 3/52 | 0.0374 |
| Positive regulation of endothelial cell apoptotic process (GO:2000353) | 2/16 | 0.0374 |
| Positive regulation of myeloid leukocyte cytokine production involved in immune response (GO:0061081) | 2/16 | 0.0374 |
| Negative regulation of gene expression (GO:0010629) | 7/322 | 0.0374 |
| Regulation of cell migration (GO:0030334) | 8/408 | 0.0375 |
| Regulation of interleukin-6 production (GO:0032675) | 4/110 | 0.0428 |
| Positive regulation of interferon gamma production (GO:0032729) | 3/57 | 0.0450 |
| Negative regulation of immune system process (GO:0002683) | 2/19 | 0.0485 |
| Tumor necrosis factor-mediated signaling pathway (GO:0033209) | 4/116 | 0.0486 |
| Positive regulation of cellular process (GO:0048522) | 10/625 | 0.0494 |
| KEGG | ||
| Autoimmune thyroid disease | 6/53 | 1.33 × 10−5 |
| Cell adhesion molecules | 7/148 | 4.01 × 10−4 |
| ABC transporters | 4/45 | 0.0014 |
| IL-17 signaling pathway | 5/94 | 0.0021 |
| Necroptosis | 5/159 | 0.0203 |
| Endocytosis | 6/252 | 0.0309 |
| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | −4.29 | 5.98 × 10−8 |
| OAS1 | 2-5-oligoadenylate synthetase 1 | −3.2 | 2.65 × 10−6 |
| OAS2 | 2-5-oligoadenylate synthetase 2 | −2.53 | 2.65 × 10−6 |
| HLA-A | major histocompatibility complex, class I, A | −2.51 | 3.08 × 10−7 |
| IRF9 | interferon regulatory factor 9 | −2.35 | 3.08× 10-7 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | −2.01 | 1.38 × 10−5 |
| IFIH1 | interferon-induced, with helicase C domain 1 | −2 | 2 × 10−4 |
| B2M | beta-2-microglobulin | −1.99 | 3.37 × 10-5 |
| DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −1.74 | 2 × 10−4 |
| USP18 | ubiquitin-specific peptidase 18 | −1.65 | 6 × 10−4 |
| OAS3 | 2-5-oligoadenylate synthetase 3 | −1.58 | 9 × 10−4 |
| HLA-F | major histocompatibility complex, class I, F | −1.56 | 2.83 × 10−6 |
| PARP14 | poly(ADP-ribose) polymerase family member 14 | −1.55 | 1.21 × 10−5 |
| HLA-DRA | major histocompatibility complex, class II, DR alpha | −1.53 | 2.1 × 10−3 |
| SAMHD1 | SAM domain and HD domain 1 | −1.5 | 9.46 × 10−5 |
| BTN3A2 | butyrophilin, subfamily 3, member A2 | −1.38 | 9.7 × 10−3 |
| HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 | −1.29 | 0.0483 |
| CFB | complement factor B | −1.15 | 0.0178 |
| EIF2AK2 | eukaryotic translation initiation factor 2-alpha kinase 2 | −1.11 | 1.9 × 10-3 |
| OASL | 2-5-oligoadenylate synthetase-like | −1.09 | 0.0204 |
| TAP2 | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | −1.09 | 0.0125 |
| ABCC3 | ATP-binding cassette subfamily C member 3 | 0.93 | 0.046 |
| ZFP36 | ZFP36 ring finger protein | 1.14 | 0.0278 |
| ABCC2 | ATP-binding cassette subfamily C member 2 | 1.33 | 8 × 10-4 |
| CLDN3 | claudin 3 | 1.42 | 4.2 × 10-3 |
| MMP13 | matrix metallopeptidase 13 | 1.93 | 5.61 × 10−6 |
| S100A7 | S100 calcium-binding protein A7 | 2.73 | 3.08 × 10−7 |
| MMP1 | matrix metallopeptidase 1 | 3.06 | 6.61 × 10−8 |
| Gene Symbol | Description | Log2 FC | FDR p-Value |
|---|---|---|---|
| IFI6 | interferon, alpha-inducible protein 6 | −4.84 | 9.24 × 10−8 |
| PSMB9 | proteasome subunit beta 9 | −2.95 | 8.75 × 10−7 |
| HLA-B | major histocompatibility complex, class I, B | −2.78 | 9.24 × 10−8 |
| IFITM1 | interferon induced transmembrane protein 1 | −2.44 | 01.58 × 10−7 |
| PSMB8 | proteasome subunit beta 8 | −2.41 | 2.52 × 10−6 |
| IFI27 | interferon, alpha-inducible protein 27 | −2.35 | 1.2 × 10−3 |
| HLA-C | major histocompatibility complex, class I, C | −1.97 | 8.86 × 10−5 |
| STAT1 | signal transducer and activator of transcription 1 | −1.94 | 3.28 × 10−6 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 | −1.56 | 0.0235 |
| CCL5 | chemokine (C-C motif) ligand 5 | −1.55 | 4 × 10−4 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | −1.42 | 2 × 10−4 |
| HLA-G | major histocompatibility complex, class I, G | −1.39 | 2 × 10−4 |
| UBE2L6 | ubiquitin-conjugating enzyme E2L 6 | −1.39 | 6.2 × 10−3 |
| ESR1 | estrogen receptor 1 | −1.38 | 0.0371 |
| TAP1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | −1.34 | 1.2 × 10−3 |
| IFITM3 | interferon induced transmembrane protein 3 | −1.24 | 8.84 × 10−5 |
| BCL2 | B-cell CLL/lymphoma 2 | −1.18 | 9.4 × 10−3 |
| PLSCR1 | phospholipid scramblase 1 | −1 | 4.6 × 10−3 |
| TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | −0.9 | 0.024 |
| HPCAL1 | hippocalcin-like 1 | 0.85 | 0.0129 |
| TNFRSF12A | tumor necrosis factor receptor superfamily, member 12A | 0.87 | 0.0216 |
| S100A6 | S100 calcium-binding protein A6 | 0.9 | 4.7 × 10−3 |
| FLNA | filamin A, alpha | 0.91 | 0.0137 |
| AHR | aryl hydrocarbon receptor | 1.05 | 0.024 |
| DUSP6 | dual specificity phosphatase 6 | 1.09 | 0.0121 |
| NR4A2 | nuclear receptor subfamily 4, group A, member 2 | 1.1 | 1.9 × 10−3 |
| FGB | fibrinogen beta chain | 1.13 | 6 × 10−4 |
| MT1X | metallothionein 1X | 1.21 | 0.0102 |
| PHLDA2 | pleckstrin homology-like domain, family A, member 2 | 1.22 | 6 × 10−4 |
| AKR1C3 | aldo-keto reductase family 1, member C3 | 1.33 | 3.1 × 10−3 |
| RGS5 | regulator of G-protein signaling 5 | 1.44 | 3 × 10−4 |
| LGALS1 | lectin, galactoside-binding, soluble, 1 | 1.48 | 1.9 × 10−3 |
| S100A9 | S100 calcium-binding protein A9 | 1.49 | 4 × 10−4 |
| HMOX1 | heme oxygenase 1 | 1.55 | 1.6 × 10−3 |
| AREG | amphiregulin | 1.61 | 6.5 × 10−3 |
| S100A8 | S100 calcium-binding protein A8 | 2.26 | 2.83 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzarito, G.; Henry, M.; Rotshteyn, T.; Leeners, B.; Dubey, R.K. Transcriptomic and Functional Evidence That miRNA193a-3p Inhibits Lymphatic Endothelial Cell (LEC) and LEC + MCF-7 Spheroid Growth Directly and by Altering MCF-7 Secretome. Cells 2023, 12, 389. https://doi.org/10.3390/cells12030389
Azzarito G, Henry M, Rotshteyn T, Leeners B, Dubey RK. Transcriptomic and Functional Evidence That miRNA193a-3p Inhibits Lymphatic Endothelial Cell (LEC) and LEC + MCF-7 Spheroid Growth Directly and by Altering MCF-7 Secretome. Cells. 2023; 12(3):389. https://doi.org/10.3390/cells12030389
Chicago/Turabian StyleAzzarito, Giovanna, Margit Henry, Tamara Rotshteyn, Brigitte Leeners, and Raghvendra K. Dubey. 2023. "Transcriptomic and Functional Evidence That miRNA193a-3p Inhibits Lymphatic Endothelial Cell (LEC) and LEC + MCF-7 Spheroid Growth Directly and by Altering MCF-7 Secretome" Cells 12, no. 3: 389. https://doi.org/10.3390/cells12030389