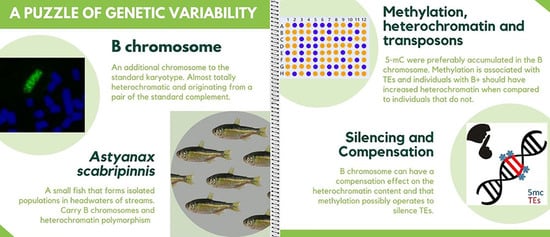
Silencing of Transposable Elements Mediated by 5-mC and Compensation of the Heterochromatin Content by Presence of B Chromosomes in Astyanax scabripinnis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Chromosomic Preparations
2.2. Quantification of the Methylation and Heterochromatic Regions
2.3. Statistical Analysis
2.4. Immunodetection of Methylated DNA, Chromosome Probe, and Fluorescence In Situ Hybridization (FISH)
2.5. Sequencing and Characterization of the Tc1-Mariner Element Obtained Fragment
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lieb, J.D.; Beck, S.; Bulyk, M.L.; Farnham, P.; Hattori, N.; Henikoff, S.; Liu, X.S.; Okumura, K.; Shiota, K.; Ushijima, T.; et al. Applying whole genome studies of epigenetic regulation to study human disease. Cytogenet. Genome Res. 2006, 114, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Vaschetto, L.M. Exploring an emerging issue: Crop epigenetics. Plant Mol. Biol. Report. 2014, 33, 751–755. [Google Scholar] [CrossRef]
- Tost, J. DNA methylation: An introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010, 44, 71–81. [Google Scholar] [CrossRef]
- Almeida-Toledo, L.F.; Viegas-Péquignot, E.; Coutinho-Barbosa, A.C.; Foresti, F.; Niveleau, A.; de Almeida Toledo-Filho, S. Localization of 5-methylcytosine in metaphase chromosomes of diploid and triploid pacu fish, Piaractus mesopotamicus (Pisces, Characiformes). Cytogenet. Genome Res. 1998, 83, 21–24. [Google Scholar] [CrossRef]
- Marques, A.; Fuchs, J.; Ma, L.; Heckmann, S.; Guerra, M.; Houben, A. Characterization of eu-and heterochromatin of Citrus with a focus on the condensation behavior of 45S rDNA chromatin. Cytogenet. Genome Res. 2011, 134, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M.; Wang, R.Y. 5-Methylcytosine in eukaryotic DNA. Science 1981, 212, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Putting the DNA back into DNA methylation. Nat. Genet. 2011, 43, 1050–1051. [Google Scholar] [CrossRef]
- Vicari, M.R.; Artoni, R.F.; Moreira-Filho, O.; Bertollo, L.A.C. Colocalization of repetitive DNA and silencing of major rRNA genes. A case report of the fish Astyanax Janeiroensis. Cytogenet. Genome Res. 2008, 122, 67–72. [Google Scholar] [CrossRef]
- Barros, A.V.; Sczepanski, T.S.; Cabrero, J.; Camacho, J.P.M.; Vicari, M.R.; Artoni, R.F. Fiber FISH reveals different patterns of highresolution physical mapping for repetitive DNA in fish. Aquaculture 2011, 322, 47–50. [Google Scholar] [CrossRef]
- Kidwell, M.G.; Lish, D.R. Perspective: Transposable elements, parasitic DNA, and genome evolution. Int. J. Org. Evol. 2001, 55, 1–24. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Alper, B.J.; Lowe, B.R.; Partridge, J.F. Centromeric heterochromatin assembly in fission yeast—balancing transcription, RNA interference and chromatin modification. Chromosome Res. 2012, 20, 521–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grewal, S.I.; Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 2003, 301, 798–802. [Google Scholar] [CrossRef] [Green Version]
- Turner, B.M. Histone acetylation and an epigenetic code. BioEssays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.N.; Rees, H. B Chromosomes; Academic Pr: London, UK, 1982; p. 266. ISBN 0123900603. [Google Scholar]
- Moreira-Filho, O.; Galetti, P.M., Jr.; Bertollo, L.A.C. B chromosomes in the fish Astyanax scabripinnis (Characidae, Tetragonopterinae): An overview in natural populations. Cytogenet. Genome Res. 2004, 106, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Vicente, V.E.; Moreira-Filho, O.; Camacho, J.P.M. Sex-ratio distortion associated with the presence of a B chromosome in Astyanax scabripinnis (Teleostei, Characidae). Cytogenet. Genome Res. 1996, 74, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Vicari, M.R.; Pistune, H.F.M.; Castro, J.P.; Almeida, M.C.; Bertollo, L.A.C.; Moreira-Filho, O.; Camacho, J.P.M.; Artoni, R.F. New insights on the origin of B chromosomes in Astyanax scabripinnis obtained by chromosome painting and FISH. Genetica 2011, 139, 1073–1081. [Google Scholar] [CrossRef]
- Mestriner, C.A.; Galetti, P.M., Jr.; Valentini, S.R.; Ruiz, I.R.G.; Abel, L.D.S.; Moreira-Filho, O.; Camacho, J.P.M. Structural and functional evidence that a B chromosome in the characid fish Astyanax scabripinnis is an isochromosome. Heredity 2000, 85, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, P.; Oliveira, L.A.; Pucci, M.B.; Santos, M.H.; Moreira-Filho, O.; Vicari, M.R.; Nogaroto, V.; Almeida, M.C.; Artoni, R.F. Identification and chromosome mapping of repetitive elements in the Astyanax scabripinnis (Teleostei: Characidae) species complex. Genetica 2015, 143, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C.; Takahashi, C.S.; Moreira-Filho, O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Braz. J. Genet. 1978, 1, 103–120. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Heredita 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffini-Castiglione, M.; Cremonini, R.; Frediani, M. DNA methylation patterns on plant chromosomes. Caryologia 2002, 55, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Schemberger, M.O.; Nogaroto, V.; Almeida, M.C.; Artoni, R.F.; Valente, G.T.; Martins, C.; Vicari, M.R. Sequence analyses and chromosomal distribution of the Tc1/Mariner element in Parodontidae fish (Teleostei: Characiformes). Gene 2016, 593, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Straume, T.; Gray, J. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Storer, J.; Hubley, R.; Rosen, J.; Wheeler, T.J.; Smit, A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. Dna 2021, 12, 2. [Google Scholar] [CrossRef]
- Chumová, Z.; Mandáková, T.; Trávníček, P. Are B-chromosomes responsible for the extraordinary genome size variation in selected Anthoxanthum annuals? Plant Syst. Evol. 2016, 302, 731–738. [Google Scholar] [CrossRef]
- Mantovani, M.; Abel, L.D.S.; Mestriner, C.A.; Moreira-Filho, O. Accentuated polymorphism of heterochromatin and nucleolar organizer regions in Astyanax scabripinnis (Pisces, Characidae): Tools for understanding karyotypic evolution. Genetica 2000, 109, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Pazza, R.; Kavalco, K.F.; Bertollo, L.A.C. Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). Chromosomal location of a satellite DNA. Cytogenet. Genome Res. 2008, 122, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Kantek, D.L.Z.; Vicari, M.R.; Peres, W.A.M.; Cestari, M.M.; Artoni, R.F.; Bertollo, L.A.C.; Moreira-Filho, O. Chromosomal location and distribution of As51 satellite DNA in five species of the genus Astyanax (Teleostei, Characidae, Incertae sedis). J. Fish Biol. 2009, 75, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, D.T.; Porto-Foresti, F. Chromosome polymorphism of heterochromatin and nucleolar regions in two populations of the fish Astyanax bockmanni (Teleostei: Characiformes). Neotrop. Ichthyol. 2010, 8, 861–866. [Google Scholar] [CrossRef]
- Wang, J.; Jia, S.T.; Jia, S. New Insights into the Regulation of Heterochromatin. Trends Genet. 2016, 32, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Lippman, Z.; Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature 2004, 431, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Steinlein, C.; Yano, C.F.; Cioffi, M.B. Hypermethylated chromosome regions in nine fish species with heteromorphic sex chromosomes. Cytogenet. Genome Res. 2015, 147, 169–178. [Google Scholar] [CrossRef]
- Bernardino, J.; Lombard, M.; Niveleau, A.; Dutrillaux, B. Common methylation characteristics of sex chromosomes in somatic and germ cells from mouse, lemur and human. Chromosome Res. 2000, 8, 513–525. [Google Scholar] [CrossRef]
- Schimid, M.; Steinlein, C. The hypermethylated regions in avian chromosomes. Cytogenet. Genome Res. 2017, 151, 216–227. [Google Scholar] [CrossRef]
- López-León, M.D.; Cabrero, J.; Camacho, J.P.M. Changes in DNA methylation during development in the B chromosome NOR of the grasshopper Eyprepocnemis plorans. Heredity 1995, 74, 296–302. [Google Scholar] [CrossRef]
- Russell, S.J.; LaMarre, J. Transposons and the PIWI pathway: Genome defense in gametes and embryos. Reproduction 2018, 156, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Bender, J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004, 55, 41–68. [Google Scholar] [CrossRef]
- Dimitri, P.; Junakovic, N.; Arcà, B. Colonization of heterochromatic genes by transposable elements in Drosophila. Mol. Biol. Evol. 2003, 20, 503–512. [Google Scholar] [CrossRef]
- Schemberger, M.O.; Oliveira, J.I.N.; Nogaroto, V.; Almeida, M.C.; Artoni, R.F.; Cestari, M.M.; Moreira-Filho, O.; Vicari, M.R. Construction and characterization of a repetitive DNA library in Parodontidae (Actinopterygii: Characiformes): A genomic and evolutionary approach to the degeneration of the W sex chromosome. Zebrafish 2014, 11, 518–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinzelle, L.; Izsvak, Z.; Ivics, Z. Molecular domestication of transposable elements: From detrimental parasites to useful host genes. Cell. Mol. Life Sci. 2009, 66, 1073–1093. [Google Scholar] [CrossRef] [PubMed]
- Koo, D.H.; Han, F.; Birchler, J.A.; Jiang, J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011, 21, 908–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Number of Cells | Mean | Lower Bound | Upper Bound | |
|---|---|---|---|---|
| 1 | 242 | 0.1273 | 0.1055 | 0.1490 |
| 2 | 233 | 0.1582 | 0.1316 | 0.1848 |
| 3 | 241 | 0.1163 | 0.0975 | 0.1352 |
| 4 | 214 | 0.1390 | 0.1137 | 0.1643 |
| Total | 930 | 0.1349 | 0.1233 | 0.1465 |
| Number of Cells | Mean | Lower Bound | Upper Bound | |
|---|---|---|---|---|
| 1 | 242 | −1.0056 | −1.0363 | −0.9749 |
| 2 | 233 | −0.9478 | −0.9857 | −0.9100 |
| 3 | 241 | −1.0385 | −1.0684 | −1.0085 |
| 4 | 214 | −1.0084 | −1.0480 | −0.9688 |
| Total | 930 | −1.0003 | −1.0176 | −0.9830 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, P.; Schemczssen-Graeff, Z.; Marques, A.; da Silva, M.; Favero, G.M.; Sobreiro, B.P.; de Almeida, M.C.; Moreira-Filho, O.; Silva, D.M.Z.d.A.; Porto-Foresti, F.; et al. Silencing of Transposable Elements Mediated by 5-mC and Compensation of the Heterochromatin Content by Presence of B Chromosomes in Astyanax scabripinnis. Cells 2021, 10, 1162. https://doi.org/10.3390/cells10051162
Barbosa P, Schemczssen-Graeff Z, Marques A, da Silva M, Favero GM, Sobreiro BP, de Almeida MC, Moreira-Filho O, Silva DMZdA, Porto-Foresti F, et al. Silencing of Transposable Elements Mediated by 5-mC and Compensation of the Heterochromatin Content by Presence of B Chromosomes in Astyanax scabripinnis. Cells. 2021; 10(5):1162. https://doi.org/10.3390/cells10051162
Chicago/Turabian StyleBarbosa, Patrícia, Zelinda Schemczssen-Graeff, André Marques, Maelin da Silva, Giovani Marino Favero, Bernardo Passos Sobreiro, Mara Cristina de Almeida, Orlando Moreira-Filho, Duílio Mazzoni Zerbinato de Andrade Silva, Fábio Porto-Foresti, and et al. 2021. "Silencing of Transposable Elements Mediated by 5-mC and Compensation of the Heterochromatin Content by Presence of B Chromosomes in Astyanax scabripinnis" Cells 10, no. 5: 1162. https://doi.org/10.3390/cells10051162