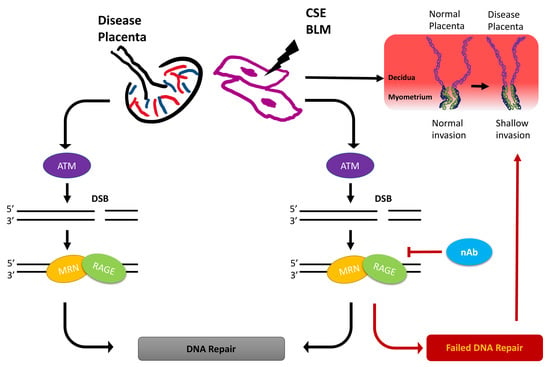
A Role for RAGE in DNA Double Strand Breaks (DSBs) Detected in Pathological Placentas and Trophoblast Cells
Abstract
:1. Introduction
2. Materials and methods
2.1. Human Placental Tissues
2.2. Cell Culture and Treatments
2.3. Cigarette Smoke Extract (CSE)
2.4. Immunofluorescence (IF)
2.5. DNA Degradation
2.6. Cytoplasmic and Nuclear Extraction
2.7. Immunoprecipitation
2.8. Western Blotting
2.9. Real Time Cell Invasion
2.10. Statistical Analysis
3. Results
3.1. RAGE and Placental DNA-DSBs
3.2. Genomic Instability in Pregnancy Complications
3.3. RAGE and Placental γ-H2AX Nuclear Expression
3.4. RAGE Interacts with ATM and MRE11 during Placental DNA-DSBs
3.5. Role of RAGE in Trophoblast DNA Damage
3.6. Trophoblast RAGE and γ-H2AX Nuclear Expression
3.7. RAGE Interacts with ATM and MRE11 during CSE and BLM Induced DNA-DSBs
3.8. Trophoblast Dysfunction as a Consequence of Genomic Instability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATM | activate ataxia-telangiectasia-mutated |
| CSE | cigarette smoke extract |
| DDR | DNA damage response |
| DSBs | double-strand breaks |
| γ-H2AX | phosphor H2A histone family member X |
| GDM | gestational diabetes mellitus |
| HMGB1 | high mobility group box 1 |
| HRR | homologous recombination repair |
| MRE11 | double-strand break repair protein MRE11 |
| MRN | MRE11-Rad50-Nbs complex |
| NHEIJ | non-homologous end joining |
| PE | preeclampsia |
| PTL | preterm labor |
| RAGE | receptors for advanced glycation end-products |
| Sw.71 | Swan 71 first trimester trophoblast cells |
| IUGR | intrauterine growth restriction |
References
- Knofler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.; James, J. Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monson, T.; Wright, T.; Galan, H.L.; Reynolds, P.R.; Arroyo, J.A. Caspase dependent and independent mechanisms of apoptosis across gestation in a sheep model of placental insufficiency and intrauterine growth restriction. Apoptosis 2017, 22, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M.; Enders, A.C.; Pijnenborg, R. The role of invasive trophoblast in implantation and placentation of primates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Damsky, C.H.; Fisher, S.J. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J. Clin. Investig. 1997, 99, 2152–2164. [Google Scholar] [CrossRef] [Green Version]
- Reuvekamp, A.; Velsing-Aarts, F.V.; Poulina, I.E.; Capello, J.J.; Duits, A.J. Selective deficit of angiogenic growth factors characterises pregnancies complicated by pre-eclampsia. Br. J. Obstet. Gynaecol. 1999, 106, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Knuth, A.; Liu, L.; Nielsen, H.; Merril, D.; Torry, D.S.; Arroyo, J.A. Placenta Growth Factor Induces Invasion and Activates p70 during Rapamycin Treatment in Trophoblast Cells. Am. J. Reprod. Immunol. 2014, 73, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Bahr, B.L.; Price, M.D.; Merrill, D.; Mejia, C.; Call, L.; Bearss, D.; Arroyo, J. Different expression of placental pyruvate kinase in normal, preeclamptic and intrauterine growth restriction pregnancies. Placenta 2014, 35, 883–890. [Google Scholar] [CrossRef]
- Arshad, R.; Karim, N.; Ara Hasan, J. Effects of insulin on placental, fetal and maternal outcomes in gestational diabetes mellitus. Pak. J. Med. Sci. 2014, 30, 240–244. [Google Scholar] [CrossRef]
- Mo, H.Q.; Tian, F.J.; Li, X.; Zhang, J.; Ma, X.L.; Zeng, W.H.; Lin, Y.; Zhang, Y. ANXA7 regulates trophoblast proliferation and apoptosis in preeclampsia. Am. J. Reprod. Immunol. 2019, 82, e13183. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Driscoll, M. Diseases associated with defective responses to DNA damage. Cold Spring Harb. Perspect. Biol. 2012, 4, a012773. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014, 19, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanna, K.K.; Lavin, M.F.; Jackson, S.P.; Mulhern, T.D. ATM, a central controller of cellular responses to DNA damage. Cell Death Differ 2001, 8, 1052–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [PubMed]
- Ramasamy, R.; Yan, S.F.; Herold, K.; Clynes, R.; Schmidt, A.M. Receptor for advanced glycation end products: Fundamental roles in the inflammatory response: Winding the way to the pathogenesis of endothelial dysfunction and atherosclerosis. Ann. N. Y. Acad. Sci. 2008, 1126, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Lewis, J.B.; Hirschi, K.M.; Arroyo, J.A.; Bikman, B.T.; Kooyman, D.L.; Reynolds, P.R. Plausible Roles for RAGE in Conditions Exacerbated by Direct and Indirect (Secondhand) Smoke Exposure. Int. J. Mol. Sci. 2017, 18, 652. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Fleming, T.; Terjung, S.; Gorzelanny, C.; Gebhardt, C.; Agrawal, R.; Mall, M.A.; Ranzinger, J.; Zeier, M.; Madhusudhan, T.; et al. Homeostatic nuclear RAGE-ATM interaction is essential for efficient DNA repair. Nucleic Acids Res. 2017, 45, 10595–10613. [Google Scholar] [CrossRef]
- Lewis, J.B.; Mejia, C.; Jordan, C.; Monson, T.D.; Bodine, J.S.; Dunaway, T.M.; Egbert, K.M.; Lewis, A.L.; Wright, T.J.; Ogden, K.C.; et al. Inhibition of the receptor for advanced glycation end-products (RAGE) protects from secondhand smoke (SHS)-induced intrauterine growth restriction IUGR in mice. Cell Tissue Res. 2017, 370, 513–521. [Google Scholar] [CrossRef]
- Furness, D.L.; Dekker, G.A.; Roberts, C.T. DNA damage and health in pregnancy. J. Reprod. Immunol. 2011, 89, 153–162. [Google Scholar] [CrossRef]
- Tomilin, N.V.; Solovjeva, L.V.; Svetlova, M.P.; Pleskach, N.M.; Zalenskaya, I.A.; Yau, P.M.; Bradbury, E.M. Visualization of focal nuclear sites of DNA repair synthesis induced by bleomycin in human cells. Radiat. Res. 2001, 156, 347–354. [Google Scholar] [CrossRef]
- Robles, S.J.; Adami, G.R. Agents that cause DNA double strand breaks lead to p16INK4a enriment and the premature senescence of normal fibroblasts. Oncogene 1998, 16, 1113–1123. [Google Scholar] [CrossRef] [Green Version]
- Argentin, G.; Cicchetti, R. Genotoxic and antiapoptotic effect of nicotine on human gingival fibroblasts. Toxicol. Sci. 2004, 79, 75–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.A.; Garcia-Jones, P.; Graham, A.; Teng, C.C.; Battaglia, F.C.; Galan, H.L. Placental TonEBP/NFAT5 osmolyte regulation in an ovine model of intrauterine growth restriction. Biol. Reprod. 2012, 86, 94. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.J.; Yang, L.X. Gamma-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar] [PubMed]
- Tadesse, S.; Kidane, D.; Guller, S.; Luo, T.; Norwitz, N.G.; Arcuri, F.; Toti, P.; Norwitz, E.R. In vivo and in vitro evidence for placental DNA damage in preeclampsia. PLoS ONE 2014, 9, e86791. [Google Scholar] [CrossRef] [Green Version]
- Slatter, T.L.; Park, L.; Anderson, K.; Lailai-Tasmania, V.; Herbison, P.; Clow, W.; Royds, J.A.; Devenish, C.; Hung, N.A. Smoking during pregnancy causes double-strand DNA break damage to the placenta. Hum. Pathol. 2014, 45, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, K.; Almasan, A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol. Biol. 2012, 920, 613–626. [Google Scholar]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.S.; Francois, M.; Fenech, M.F.; Leifert, W.R. Persistent gammaH2AX: A promising molecular marker of DNA damage and aging. Mutat. Res. Rev. Mutat. Res. 2015, 766, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Barone, F.C.; Irving, E.A.; Ray, A.M.; Lee, J.C.; Kassis, S.; Kumar, S.; Badger, A.M.; Legos, J.J.; Erhardt, J.A.; Ohlstein, E.H.; et al. Inhibition of p38 mitogen-activated protein kinase provides neuroprotection in cerebral focal ischemia. Med. Res. Rev. 2001, 21, 129–145. [Google Scholar] [CrossRef]
- Lavin, M.F.; Kozlov, S.; Gatei, M.; Kijas, A.W. ATM-Dependent Phosphorylation of All Three Members of the MRN Complex: From Sensor to Adaptor. Biomolecules 2015, 5, 2877–2902. [Google Scholar] [CrossRef] [Green Version]
- Lavin, M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 2007, 26, 7749–7758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [Green Version]









| Control | GDM | PTL | PE | P Value | |
|---|---|---|---|---|---|
| Maternal Age | 34 ± 2.96 | 35 ± 2.8 | 29 ± 1.6 | 36 ± 2.2 | 0.494 |
| Gestational Age (wks) | 38 ± 0.02 | 39 ± 1.6 | 32 ± 0.5 | 32 ± 2.3 | 0.002 |
| Fetal Weight (g) | 3498 ± 59 | 3247 ± 172 | 2025 ± 139 | 2025 ± 139 | 0.002 |
| Antibody | Species | Supplier | Application |
|---|---|---|---|
| RAGE | Goat | R&D (AF1145) | WB, IF |
| RAGE | Mouse | Abcam (ab89911) | Neutralizing |
| Phospho-γ-H2AX | Rabbit | Cell Signaling (9718) | WB, IF |
| Phospho-pATM | Rabbit | Abcam (ab81292) | IP |
| MRE11 | Rabbit | Cell Signaling (4895) | IP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, K.Y.F.; Tullis, B.; Breithaupt, K.L.; Fowers, R.; Jones, N.; Grajeda, S.; Reynolds, P.R.; Arroyo, J.A. A Role for RAGE in DNA Double Strand Breaks (DSBs) Detected in Pathological Placentas and Trophoblast Cells. Cells 2021, 10, 857. https://doi.org/10.3390/cells10040857
Tsai KYF, Tullis B, Breithaupt KL, Fowers R, Jones N, Grajeda S, Reynolds PR, Arroyo JA. A Role for RAGE in DNA Double Strand Breaks (DSBs) Detected in Pathological Placentas and Trophoblast Cells. Cells. 2021; 10(4):857. https://doi.org/10.3390/cells10040857
Chicago/Turabian StyleTsai, Kary Y.F., Benton Tullis, Katrina L. Breithaupt, Rylan Fowers, Nelson Jones, Samuel Grajeda, Paul R. Reynolds, and Juan A. Arroyo. 2021. "A Role for RAGE in DNA Double Strand Breaks (DSBs) Detected in Pathological Placentas and Trophoblast Cells" Cells 10, no. 4: 857. https://doi.org/10.3390/cells10040857