The F-Actin-Binding MPRIP Forms Phase-Separated Condensates and Associates with PI(4,5)P2 and Active RNA Polymerase II in the Cell Nucleus
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Cultures and Transfections
2.2. Constructs and Antibodies
- Anti-MPRIP antibody-HPA022901 (SigmaAldrich, St. Louis, MO, USA)
- Anti-PI (4,5) P2 antibody: Z-A045, clone 2C11 (Echelon, San Jose, CA, USA)
- Anti-RNAPII CTD Phospho S5 antibody-ab5131 (Abcam, Cambridge, UK)
- Anti-Lamin B1 antibody—ab16048 (Abcam, Cambridge, UK)
- Anti-GAPDH antibody [6C5]-ab8245 (Abcam, Cambridge, UK)
- Rabbit Anti-Mouse Immunoglobulin G H&L—ab46540 (Abcam, Cambridge, UK)
- Anti-MYO1C antibody was supplied by Peter G. Gillespie, Oregon Hearing Research Center and Vollum Institute [29].
2.3. Bioinformatics Analyses
2.4. Immunofluorescence Labelling
2.5. Stimulated Emission Depletion (STED) Microscopy
2.6. Nuclear Extraction and Pull-Down Assay
2.7. Co-Immunoprecipitation Assay
2.8. Live-Cell Imaging Microscopy
2.9. Hexanediol Treatment
2.10. FRAP
2.11. Phalloidin Staining of GFP-MPRIP Expressing U2OS Cells
3. Results
3.1. MPRIP Protein Is Present in the Cell Nucleus
3.2. Nuclear MPRIP Interacts with PIP2 and Forms a Complex with RNAPII and MYO1C
3.3. Overexpression of MPRIP Leads to Formation of Condensates and Fibrous Structures in the Cell Nucleus
3.4. Nuclear MPRIP Condensates Are Formed by Phase Separation
3.5. Dynamic Liquid-Like MPRIP Condensates Are Able to Fuse, Segregate and Form Fibers
3.6. MPRIP Condensates Show High Internal Dynamics and Rapid Molecular Interchange between Condensates and Neighboring Nucleoplasm
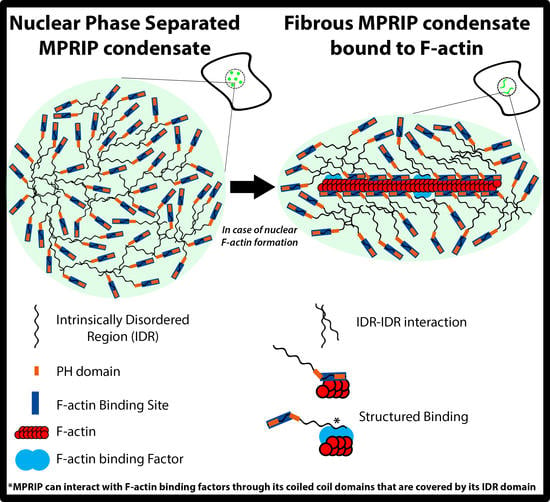
3.7. MPRIP Fibers Contain Nuclear F-Actin
3.8. The C-Terminal IDR Domain Is Responsible for Phase Separation of MPRIP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mulder, J.; Poland, M.; Gebbink, M.F.; Calafat, J.; Moolenaar, W.H.; Kranenburg, O. p116Rip is a novel filamentous actin-binding protein. J. Biol. Chem. 2003, 278, 27216–27223. [Google Scholar] [CrossRef] [Green Version]
- Vallenius, T.; Vaahtomeri, K.; Kovac, B.; Osiceanu, A.M.; Viljanen, M.; Makela, T.P. An association between NUAK2 and MRIP reveals a novel mechanism for regulation of actin stress fibers. J. Cell Sci. 2011, 124 Pt 3, 384–393. [Google Scholar] [CrossRef] [Green Version]
- Koga, Y.; Ikebe, M. p116Rip Decreases Myosin II Phosphorylation by Activating Myosin Light Chain Phosphatase and by Inactivating RhoA. J. Biol. Chem. 2005, 280, 4983–4991. [Google Scholar] [CrossRef] [Green Version]
- Sztacho, M.; Salovska, B.; Cervenka, J.; Balaban, C.; Hoboth, P.; Hozak, P. Limited Proteolysis-Coupled Mass Spectrometry Identifies Phosphatidylinositol 4,5-Bisphosphate Effectors in Human Nuclear Proteome. Cells 2021, 10, 68. [Google Scholar] [CrossRef]
- Mulder, J.; Ariaens, A.; van den Boomen, D.; Moolenaar, W.H. p116Rip targets myosin phosphatase to the actin cytoskeleton and is essential for RhoA/ROCK-regulated neuritogenesis. Mol. Biol. Cell 2004, 15, 5516–5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, J.; Ariaens, A.; van Horck, F.P.G.; Moolenaar, W.H. Inhibition of RhoA-mediated SRF activation by p116Rip. FEBS Lett. 2005, 579, 6121–6127. [Google Scholar] [CrossRef] [Green Version]
- Surks, H.K.; Riddick, N.; Ohtani, K. M-RIP Targets Myosin Phosphatase to Stress Fibers to Regulate Myosin Light Chain Phosphorylation in Vascular Smooth Muscle Cells. J. Biol. Chem. 2005, 280, 42543–42551. [Google Scholar] [CrossRef] [Green Version]
- Surks, H.K.; Richards, C.T.; Mendelsohn, M.E. Myosin Phosphatase-Rho Interacting Protein a New Member of the Myosin Phosphatase Complex that Directly Binds RhoA. J. Biol. Chem. 2003, 278, 51484–51493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, E.; Kalli, A.C.; Yasuoka, K.; Sansom, M.S.P. Interactions of Pleckstrin Homology Domains with Membranes: Adding Back the Bilayer via High-Throughput Molecular Dynamics. Structure 2016, 24, 1421–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almuzzaini, B.; Sarshad, A.A.; Farrants, A.K.; Percipalle, P. Nuclear myosin 1 contributes to a chromatin landscape compatible with RNA polymerase II transcription activation. BMC Biol. 2015, 13, 35. [Google Scholar] [CrossRef] [Green Version]
- Nevzorov, I.; Sidorenko, E.; Wang, W.; Zhao, H.; Vartiainen, M.K. Myosin-1C uses a novel phosphoinositide-dependent pathway for nuclear localization. EMBO Rep. 2018, 19, 290–304. [Google Scholar] [CrossRef]
- Sobol, M.; Krausova, A.; Yildirim, S.; Kalasova, I.; Faberova, V.; Vrkoslav, V.; Philimonenko, V.; Marasek, P.; Pastorek, L.; Capek, M.; et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Castano, E.; Yildirim, S.; Fáberová, V.; Krausová, A.; Uličná, L.; Paprčková, D.; Sztacho, M.; Hozák, P. Nuclear Phosphoinositides—Versatile Regulators of Genome Functions. Cells 2019, 8, 649. [Google Scholar] [CrossRef] [Green Version]
- Sztacho, M.; Sobol, M.; Balaban, C.; Escudeiro Lopes, S.E.; Hozak, P. Nuclear phosphoinositides and phase separation: Important players in nuclear compartmentalization. Adv. Biol. Regul. 2019, 71, 111–117. [Google Scholar] [CrossRef]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar sub-compartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, I.A.; Bartek, J.; Dundr, M. Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin. Cell Dev. Biol. 2019, 90, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Brangwynne, C.P. The liquid nucleome–phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, jcs235093. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Chen, X.; Wu, X.; Zhang, M. Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J. Biol. Chem. 2019, 294, 14823–14835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [Green Version]
- Nowak, G.; Pestic-Dragovich, L.; Hozak, P.; Philimonenko, A.; Simerly, C.; Schatten, G.; de Lanerolle, P. Evidence for the presence of myosin I in the nucleus. J. Biol. Chem. 1997, 272, 17176–17181. [Google Scholar] [CrossRef] [Green Version]
- Pestic-Dragovich, L.; Stojiljkovic, L.; Philimonenko, A.A.; Nowak, G.; Ke, Y.; Settlage, R.E.; Shabanowitz, J.; Hunt, D.F.; Hozak, P.; de Lanerolle, P. A myosin I isoform in the nucleus. Science 2000, 290, 337–341. [Google Scholar] [CrossRef]
- Dumont, R.A.; Zhao, Y.D.; Holt, J.R.; Bahler, M.; Gillespie, P.G. Myosin-I isozymes in neonatal rodent auditory and vestibular epithelia. J. Assoc. Res. Otolaryngol. 2002, 3, 375–389. [Google Scholar] [CrossRef] [Green Version]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008, 183, 223–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, Z.H.; Jones, D.R.; Sommer, L.; Foulger, R.; Bultsma, Y.; D’Santos, C.; Divecha, N. Nuclear phosphoinositides and their impact on nuclear functions. FEBS J. 2013, 280, 6295–6310. [Google Scholar] [CrossRef]
- Yildirim, S.; Castano, E.; Sobol, M.; Philimonenko, V.V.; Dzijak, R.; Venit, T.; Hozak, P. Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J. Cell Sci. 2013, 126 Pt 12, 2730–2739. [Google Scholar] [CrossRef] [Green Version]
- Kroschwald, S.; Maharana, S.; Mateju, D.; Malinovska, L.; Nüske, E.; Poser, I.; Richter, D.; Alberti, S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 2015, 4, e06807. [Google Scholar] [CrossRef]
- Patel, S.S.; Belmont, B.J.; Sante, J.M.; Rexach, M.F. Natively Unfolded Nucleoporins Gate Protein Diffusion across the Nuclear Pore Complex. Cell 2007, 129, 83–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; McKnight, S.L. A Solid-State Conceptualization of Information Transfer from Gene to Message to Protein. Annu. Rev. Biochem. 2018, 87, 351–390. [Google Scholar] [CrossRef]
- Nair, S.J.; Yang, L.; Meluzzi, D.; Oh, S.; Yang, F.; Friedman, M.J.; Wang, S.; Suter, T.; Alshareedah, I.; Gamliel, A.; et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019, 26, 193–203. [Google Scholar] [CrossRef]
- Lin, Y.; Mori, E.; Kato, M.; Xiang, S.; Wu, L.; Kwon, I.; McKnight, S.L. Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell 2016, 167, 789–802.e12. [Google Scholar] [CrossRef] [Green Version]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P Granules Are Liquid Droplets that Localize by Controlled Dissolution/Condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Yamamoto, M.; Hilgemann, D.H.; Feng, S.; Bito, H.; Ishihara, H.; Shibasaki, Y.; Yin, H.L. Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J. Cell Biol. 2001, 152, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Shibasaki, Y.; Ishihara, H.; Kizuki, N.; Asano, T.; Oka, Y.; Yazaki, Y. Massive actin polymerization induced by phosphatidylinositol-4-phosphate 5-kinase in vivo. J. Biol. Chem. 1997, 272, 7578–7581. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Ma, L.; Parrini, M.C.; Mao, X.; Lopez, M.; Wu, C.; Marks, P.W.; Davidson, L.; Kwiatkowski, D.J.; Kirchhausen, T.; et al. Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr. Biol. 2000, 10, 758–765. [Google Scholar] [CrossRef] [Green Version]
- Tsujita, K.; Itoh, T. Phosphoinositides in the regulation of actin cortex and cell migration. Biochim. Biophys. Acta 2015, 1851, 824–831. [Google Scholar] [CrossRef] [Green Version]
- Riggi, M.; Niewola-Staszkowska, K.; Chiaruttini, N.; Colom, A.; Kusmider, B.; Mercier, V.; Soleimanpour, S.; Stahl, M.; Matile, S.; Roux, A.; et al. Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nat. Cell Biol. 2018, 20, 1043–1051. [Google Scholar] [CrossRef]
- Sztacho, M.; Segeletz, S.; Sanchez-Fernandez, M.A.; Czupalla, C.; Niehage, C.; Hoflack, B. BAR Proteins PSTPIP1/2 Regulate Podosome Dynamics and the Resorption Activity of Osteoclasts. PLoS ONE 2016, 11, e0164829. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Shiraishi, M.; Fukami, K.; Tanabe, A.; Ikeda-Matsuo, Y.; Naito, Y.; Sasaki, Y. MARCKS regulates lamellipodia formation induced by IGF-I via association with PIP2 and beta-actin at membrane microdomains. J. Cell. Physiol. 2009, 220, 748–755. [Google Scholar] [CrossRef]
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marášek, P.; Castaño, E.; Hozák, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486. [Google Scholar] [CrossRef] [Green Version]
- Ulicna, L.; Kalendova, A.; Kalasova, I.; Vacik, T.; Hozák, P. PIP2 epigenetically represses rRNA genes transcription interacting with PHF8. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Bavelloni, A.; Faenza, I.; Cioffi, G.; Piazzi, M.; Parisi, D.; Matic, I.; Maraldi, N.M.; Cocco, L. Proteomic-based analysis of nuclear signaling: PLCbeta1 affects the expression of the splicing factor SRp20 in Friend erythroleukemia cells. Proteomics 2006, 6, 5725–5734. [Google Scholar] [CrossRef]
- Faenza, I.; Ramazzotti, G.; Bavelloni, A.; Fiume, R.; Gaboardi, G.C.; Follo, M.Y.; Gilmour, R.S.; Martelli, A.M.; Ravid, K.; Cocco, L. Inositide-dependent phospholipase C signaling mimics insulin in skeletal muscle differentiation by affecting specific regions of the cyclin D3 promoter. Endocrinology 2007, 148, 1108–1117. [Google Scholar] [CrossRef] [Green Version]
- Lewis, A.E.; Sommer, L.; Arntzen, M.O.; Strahm, Y.; Morrice, N.A.; Divecha, N.; D’Santos, C.S. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell Proteomics 2011, 10, M110.003376. [Google Scholar] [CrossRef] [Green Version]
- Hoboth, P.; Sztacho, M.; Sebesta, O.; Schatz, M.; Castano, E.; Hozak, P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 158890. [Google Scholar] [CrossRef]
- Hokanson, D.E.; Laakso, J.M.; Lin, T.; Sept, D.; Ostap, E.M. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell 2006, 17, 4856–4865. [Google Scholar] [CrossRef] [Green Version]
- Hokanson, D.E.; Ostap, E.M. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc. Nat. Acad. Sci. USA 2006, 103, 3118–3123. [Google Scholar] [CrossRef] [Green Version]
- Philimonenko, V.V.; Zhao, J.; Iben, S.; Dingova, H.; Kysela, K.; Kahle, M.; Zentgraf, H.; Hofmann, W.A.; de Lanerolle, P.; Hozak, P.; et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004, 6, 1165–1172. [Google Scholar] [CrossRef]
- Ye, J.; Zhao, J.; Hoffmann-Rohrer, U.; Grummt, I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008, 22, 322–330. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Reed, R. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Nat. Acad. Sci. USA 2015, 112, 8608–8613. [Google Scholar] [CrossRef] [Green Version]
- Kristo, I.; Bajusz, C.; Borsos, B.N.; Pankotai, T.; Dopie, J.; Jankovics, F.; Vartiainen, M.K.; Erdelyi, M.; Vilmos, P. The actin binding cytoskeletal protein Moesin is involved in nuclear mRNA export. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1589–1604. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, M.; Moore, H.M.; Prajapati, B.; Dopie, J.; Merilainen, L.; Honkanen, M.; Matos, R.C.; Poukkula, M.; Hietakangas, V.; Vartiainen, M.K. Nuclear Actin Is Required for Transcription during Drosophila Oogenesis. iScience 2018, 9, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Viita, T.; Kyheroinen, S.; Prajapati, B.; Virtanen, J.; Frilander, M.J.; Varjosalo, M.; Vartiainen, M.K. Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venit, T.; Semesta, K.; Farrukh, S.; Endara-Coll, M.; Havalda, R.; Hozak, P.; Percipalle, P. Nuclear myosin 1 activates p21 gene transcription in response to DNA damage through a chromatin-based mechanism. Commun. Biol. 2020, 3, 115. [Google Scholar] [CrossRef] [PubMed]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef]
- Hurst, V.; Shimada, K.; Gasser, S.M. Nuclear Actin and Actin-Binding Proteins in DNA Repair. Trends Cell Biol. 2019, 29, 462–476. [Google Scholar] [CrossRef] [Green Version]
- Serebryannyy, L.A.; Parilla, M.; Annibale, P.; Cruz, C.M.; Laster, K.; Gratton, E.; Kudryashov, D.; Kosak, S.T.; Gottardi, C.J.; de Lanerolle, P. Persistent nuclear actin filaments inhibit transcription by RNA polymerase II. J. Cell Sci. 2016, 129, 3412–3425. [Google Scholar] [CrossRef] [Green Version]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaban, C.; Sztacho, M.; Blažíková, M.; Hozák, P. The F-Actin-Binding MPRIP Forms Phase-Separated Condensates and Associates with PI(4,5)P2 and Active RNA Polymerase II in the Cell Nucleus. Cells 2021, 10, 848. https://doi.org/10.3390/cells10040848
Balaban C, Sztacho M, Blažíková M, Hozák P. The F-Actin-Binding MPRIP Forms Phase-Separated Condensates and Associates with PI(4,5)P2 and Active RNA Polymerase II in the Cell Nucleus. Cells. 2021; 10(4):848. https://doi.org/10.3390/cells10040848
Chicago/Turabian StyleBalaban, Can, Martin Sztacho, Michaela Blažíková, and Pavel Hozák. 2021. "The F-Actin-Binding MPRIP Forms Phase-Separated Condensates and Associates with PI(4,5)P2 and Active RNA Polymerase II in the Cell Nucleus" Cells 10, no. 4: 848. https://doi.org/10.3390/cells10040848