Macrophage Resistance to Ionizing Radiation Exposure Is Accompanied by Decreased Cathepsin D and Increased Transferrin Receptor 1 Expression
Abstract
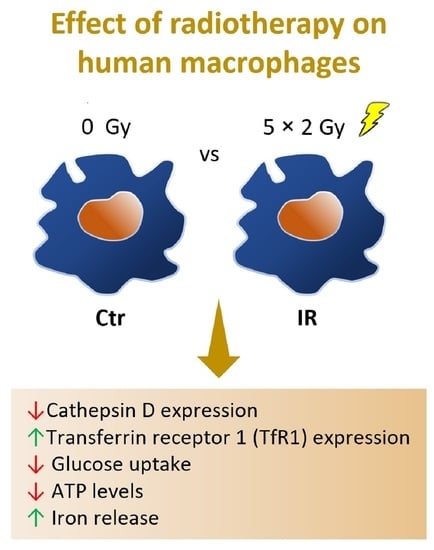
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Macrophage Culture and Exposure to IR
2.2. Cell Metabolic Activity
2.3. Glucose Uptake
2.4. Lactate Levels
2.5. Total Levels of Cellular ATP
2.6. Iron Supplementation
2.7. Iron Quantification
2.8. Proteomics
2.9. Western Blot Analysis
2.10. Animal Experiments
2.11. Immunohistochemistry for TfR1
2.12. Statistical Analysis
3. Results
3.1. Despite DNA Damage, Irradiated Macrophages Remain Viable
3.2. IR Interferes with Macrophage Metabolism and Regulation of Transport
3.3. Cathepsin D, a Positive Regulator of Apoptosis, Is Downregulated in Irradiated Macrophages
3.4. Irradiated Macrophages Exhibit a Reduction of Both Glucose Uptake and Total ATP Levels
3.5. Transferrin Receptor Protein 1 (TfR1 or CD71), a Receptor Specialized in Cellular Iron Uptake, Is Upregulated in Irradiated Macrophages
3.6. Macrophage Iron Metabolism Is Affected by IR Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, B.J.; Bezak, E.; Marcu, L.G. Quo Vadis Radiotherapy? Technological Advances and the Rising Problems in Cancer Management. BioMed Res. Int. 2013, 2013, 749203. [Google Scholar] [CrossRef] [PubMed]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate Partners for Tumor Cell Migration, Invasion, and Metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Palma, M.; Lewis, C.E. Macrophage Regulation of Tumor Responses to Anticancer Therapies. Cancer Cell 2013, 23, 277–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Esaulova, E.; Ward, J.P.; Malkova, O.N.; Runci, D.; Wong, P.; Noguchi, T.; Arthur, C.D.; Meng, W.; Alspach, E.; et al. High-Dimensional Analysis Delineates Myeloid and Lymphoid Compartment Remodeling during Successful Immune-Checkpoint Cancer Therapy. Cell 2018, 175, 1014–1030.e19. [Google Scholar] [CrossRef] [Green Version]
- McBride, W.H.; Chiang, C.-S.; Olson, J.L.; Wang, C.-C.; Hong, J.-H.; Pajonk, F.; Dougherty, G.J.; Iwamoto, K.S.; Pervan, M.; Liao, Y.-P. A Sense of Danger from Radiation. Radiat. Res. 2004, 162, 1–19. [Google Scholar] [CrossRef]
- Lauber, K.; Ernst, A.; Orth, M.; Herrmann, M.J.; Belka, C. Dying cell clearance and its impact on the outcome of tumor radiotherapy. Front. Oncol. 2012, 2, 116. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Allouch, A.; Martins, I.; Modjtahedi, N.; Deutsch, E.; Perfettini, J.-L. Macrophage biology plays a central role during ionizing radiation-elicited tumor response. Biomed. J. 2017, 40, 200–211. [Google Scholar] [CrossRef]
- Barker, H.E.; Paget, J.T.E.; Khan, A.; Harrington, K. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef] [Green Version]
- Heylmann, D.; Rödel, F.; Kindler, T.; Kaina, B. Radiation sensitivity of human and murine peripheral blood lymphocytes, stem and progenitor cells. Biochim. Biophys. Acta 2014, 1846, 121–129. [Google Scholar] [CrossRef]
- Olwell, P.M.; Cottell, D.C.; Shúilleabháin, S.N.; Maderna, P.; Seymour, C.; Mothersill, C.; Lyng, F.M. Cytoskeletal Reorganization and Altered Phagocytotic Ability in Primary Cultures of Rainbow Trout Hemopoietic Tissue Exposed to Low-Level Ionizing Radiation. Radiat. Res. 2005, 164, 45–52. [Google Scholar] [CrossRef]
- Liu, S.-Z.; Jin, S.-Z.; Liu, X.-D.; Sun, Y.-M. Role of CD28/B7 costimulation and IL-12/IL-10 interaction in the radiation-induced immune changes. BMC Immunol. 2001, 2, 8. [Google Scholar] [CrossRef]
- Xie, C.H.; Zhou, Y.; Xia, L.; He, Z.S.; Ouyang, W.; Xie, Z.H. Modulation of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 in RAW264.7 cells by irradiation. Mol. Med. Rep. 2010, 3, 809–813. [Google Scholar] [CrossRef] [Green Version]
- Conrad, S.; Ritter, S.; Fournier, C.; Nixdorff, K. Differential Effects of Irradiation with Carbon Ions and X-Rays on Macrophage Function. J. Radiat. Res. 2009, 50, 223–231. [Google Scholar] [CrossRef]
- Ibuki, Y.; Mizuno, S.; Goto, R. γ-Irradiation-induced DNA damage enhances NO production via NF-κB activation in RAW264.7 cells. Biochim. et Biophys. Acta 2003, 1593, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Shankar, B.S.; Sharma, D.; Sainis, K.B. Low dose radiation induced immunomodulation: Effect on macrophages and CD8+ T cells. Int. J. Radiat. Biol. 2005, 81, 801–812. [Google Scholar] [CrossRef]
- Tsukimoto, M.; Homma, T.; Mutou, Y.; Kojima, S. 0.5 Gy Gamma Radiation Suppresses Production of TNF-α through Up-regulation of MKP-1 in Mouse Macrophage RAW264.7 Cells. Radiat. Res. 2009, 171, 219–224. [Google Scholar] [CrossRef]
- Frischholz, B.; Wunderlich, R.; Rühle, P.-F.; Schorn, C.; Rödel, F.; Keilholz, L.; Fietkau, R.; Gaipl, U.S.; Frey, B. Reduced secretion of the inflammatory cytokine IL-1β by stimulated peritoneal macrophages of radiosensitive Balb/c mice after exposure to 0.5 or 0.7Gy of ionizing radiation. Autoimmunity 2013, 46, 323–328. [Google Scholar] [CrossRef]
- Lödermann, B.; Wunderlich, R.; Frey, S.; Schorn, C.; Stangl, S.; Rödel, F.; Keilholz, L.; Fietkau, R.; Gaipl, U.S.; Frey, B. Low dose ionising radiation leads to a NF-κB dependent decreased secretion of active IL-1β by activated macrophages with a discontinuous dose-dependency. Int. J. Radiat. Biol. 2012, 88, 727–734. [Google Scholar] [CrossRef]
- United Nations Scientific Committee on the Effects of Atomic Radiation (UNSCEAR). Biological Mechanisms of Radiation Actions at Low Doses. A White Paper to Guide the Scientific Committee’s Future Programme of Work; UNSCEAR: New York, NY, USA, 2012. [Google Scholar]
- Hellevik, T.; Martinez-Zubiaurre, I. Radiotherapy and the Tumor Stroma: The Importance of Dose and Fractionation. Front. Oncol. 2014, 4, 1. [Google Scholar] [CrossRef]
- Paiella, S.; Malleo, G.; Simoni, N.; Micera, R.; Guariglia, S.; Cavedon, C.; Marchegiani, G.; Esposito, A.; Landoni, L.; Casetti, L.; et al. A phase II trial proposal of total neoadjuvant treatment with primary chemotherapy, stereotactic body radiation therapy, and intraoperative radiation therapy in borderline resectable pancreatic adenocarcinoma. BMC Cancer 2021, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.T.; Pinto, M.L.; Cardoso, A.P.; Monteiro, C.; Maia, A.F.; Castro, P.; Figueira, R.; Monteiro, A.; Marques, M.; Mareel, M.; et al. Ionizing radiation modulates human macrophages towards a pro-inflammatory phenotype preserving their pro-invasive and pro-angiogenic capacities. Sci. Rep. 2016, 6, 18765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, A.T.; Pinto, M.L.; Velho, S.; Pinto, M.T.; Cardoso, A.P.; Figueira, R.; Monteiro, A.; Marques, M.; Seruca, R.; Barbosa, M.A.; et al. Intricate Macrophage-Colorectal Cancer Cell Communication in Response to Radiation. PLoS ONE 2016, 11, e0160891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimzadeh, O.; Atkinson, M.J.; Tapio, S. Proteomics in radiation research: Present status and future perspectives. Radiat. Environ. Biophys. 2014, 53, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Stulik, J.; Koupilová, K.; Hernychová, L.; Macela, A.; Bláha, V.; Baaske, C.; Kaffenberger, W.; Van Beuningen, D. Modulation of signal transduction pathways and global protein composition of macrophages by ionizing radiation. Electrophoresis 1999, 20, 962–968. [Google Scholar] [CrossRef]
- Chen, C.; Boylan, M.T.; Evans, C.A.; Whetton, A.A.D.; Wright, E.G. Application of Two-Dimensional Difference Gel Electrophoresis to Studying Bone Marrow Macrophages and Their in Vivo Responses to Ionizing Radiation. J. Proteome Res. 2005, 4, 1371–1380. [Google Scholar] [CrossRef]
- Smallwood, H.S.; Lopez-Ferrer, D.; Eberlein, P.E.; Watson, D.J.; Squier, T.C. Calmodulin Mediates DNA Repair Pathways Involving H2AX in Response to Low-Dose Radiation Exposure of RAW 264.7 Macrophages. Chem. Res. Toxicol. 2009, 22, 460–470. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C.W. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Bandow, J.; Sherman, J.; Baker, J.D.; Brown, P.W.; McDowell, M.T.; Molloy, M. iTRAQ Experimental Design for Plasma Biomarker Discovery. J. Proteome Res. 2008, 7, 2952–2958. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mosteller, F.; Fisher, R.A. Questions and Answers. Am. Stat. 1948, 2, 30–31. [Google Scholar] [CrossRef]
- Fishe, R. Statistical Methods, Experimental Design, and Scientific Inference: A Re-Issue of Statistical Methods for Research Workers, The Design of Experiments, and Statistical Methods and Scientific Inference; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Castro, F.; Pinto, M.L.; Pereira, C.L.; Serre, K.; Barbosa, M.A.; Vermaelen, K.; Gärtner, F.; Gonçalves, R.M.; De Wever, O.; Oliveira, M.J. Chitosan/γ-PGA nanoparticles-based immunotherapy as adjuvant to radiotherapy in breast cancer. Biomaterials 2020, 257, 120218. [Google Scholar] [CrossRef]
- Vieira, O.; Botelho, R.; Grinstein, S. Phagosome maturation: Aging gracefully. Biochem. J. 2002, 366 Pt 3, 689–704. [Google Scholar] [CrossRef]
- Minarowska, A.; Gacko, M.; Karwowska, A.; Minarowski, L. Human cathepsin D. Folia Histochem. Cytobiol. 2008, 46, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Rijnboutt, S.; Stoorvogel, W.; Geuze, H.J.; Strous, G.J. Identification of subcellular compartments involved in biosynthetic processing of cathepsin D. J. Biol. Chem. 1992, 267, 15665–15672. [Google Scholar] [CrossRef]
- Zaidi, N.; Maurer, A.; Nieke, S.; Kalbacher, H. Cathepsin D: A cellular roadmap. Biochem. Biophys. Res. Commun. 2008, 376, 5–9. [Google Scholar] [CrossRef]
- Grant, G.A. D-3-Phosphoglycerate Dehydrogenase. Front. Mol. Biosci. 2018, 5, 110. [Google Scholar] [CrossRef]
- Augustin, W.; Trümper, L.; Spengler, V. Metabolism of intact reticulocytes and mitochondria under lowered energy load induced by cycloheximide. Biomed. Biochim. Acta 1987, 46, S229–S233. [Google Scholar] [PubMed]
- Bernstein, B.W.; Bamburg, J.R. Actin-ATP Hydrolysis Is a Major Energy Drain for Neurons. J. Neurosci. 2003, 23, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Bonora, M.; Patergnani, S.; Rimessi, A.; De Marchi, E.; Suski, J.M.; Bononi, A.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. ATP synthesis and storage. Purinergic Signal. 2012, 8, 343–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodish, H.F.; Berk, A.; Darnell, J.E.; Zipursky, S.L.; Baltimore, D.; Darnell, J. Section 15.5 Active Transport by ATP-Powered Pumps. In Molecular Cell Biology, 4th ed.; W. H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Mertens, C.; Marques, O.; Horvat, N.; Simonetti, M.; Muckenthaler, M.; Jung, M. The Macrophage Iron Signature in Health and Disease. Int. J. Mol. Sci. 2021, 22, 8457. [Google Scholar] [CrossRef] [PubMed]
- Recalcati, S.; Cairo, G. Macrophages and Iron: A Special Relationship. Biomedicines 2021, 9, 1585. [Google Scholar] [CrossRef]
- Pach, R.; Sierzega, M.; Szczepanik, A.; Popiela, T.; Richter, P. Preoperative radiotherapy 5 × 5 Gy and short versus long interval between surgery for resectable rectal cancer: 10-Year follow-up of the randomised controlled trial. Radiother. Oncol. 2021, 164, 268–274. [Google Scholar] [CrossRef]
- Aderem, A.; Underhill, D.M. MECHANISMS OF PHAGOCYTOSIS IN MACROPHAGES. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef]
- Kato, T.; Kojima, K.; Murachi, T. Proteases of macrophages in rat peritoneal exudate, with special reference to the effects of actinomycete protease inhibitors. Biochim. Biophys. Acta 1972, 289, 187–193. [Google Scholar] [CrossRef]
- Benes, P.; Vetvicka, V.; Fusek, M. Cathepsin D—Many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008, 68, 12–28. [Google Scholar] [CrossRef] [Green Version]
- Boya, P.; Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 2008, 27, 6434–6451. [Google Scholar] [CrossRef]
- Appelqvist, H.; Johansson, A.C.; Linderoth, E.; Johansson, U.; Antonsson, B.; Steinfeld, R.; Kågedal, K.; Ollinger, K. Lysosome-mediated apoptosis is associated with cathepsin D-specific processing of bid at Phe24, Trp48, and Phe183. Ann. Clin. Lab. Sci. 2012, 42, 231–242. [Google Scholar]
- Bossy-Wetzel, E.; Newmeyer, D.D.; Green, D. Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J. 1998, 17, 37–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewley, M.A.; Pham, T.K.; Marriott, H.; Noirel, J.; Chu, H.-P.; Ow, S.Y.; Ryazanov, A.G.; Read, R.; Whyte, M.K.B.; Chain, B.; et al. Proteomic Evaluation and Validation of Cathepsin D Regulated Proteins in Macrophages Exposed to Streptococcus pneumoniae. Mol. Cell. Proteom. 2011, 10, M111.008193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.-H.; Jung, S.-Y.; Ahn, J.; Hwang, S.-G.; Woo, H.-J.; An, S.; Nam, S.Y.; Lim, D.-S.; Song, J.-Y. Quantitative proteomic analysis of single or fractionated radiation-induced proteins in human breast cancer MDA-MB-231 cells. Cell Biosci. 2015, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Raja, S.M.; Zazzeroni, F.; Metkar, S.S.; Shah, R.; Zhang, M.; Wang, Y.; Brömme, D.; Russin, W.A.; Lee, J.C.; et al. NF-κB protects from the lysosomal pathway of cell death. EMBO J. 2003, 22, 5313–5322. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Belguise, K.; O’Neill, C.F.; Sánchez-Morgan, N.; Romagnoli, M.; Eddy, S.F.; Mineva, N.D.; Yu, Z.; Min, C.; Trinkaus-Randall, V.; et al. RelB NF-κB Represses Estrogen Receptor α Expression via Induction of the Zinc Finger Protein Blimp1. Mol. Cell. Biol. 2009, 29, 3832–3844. [Google Scholar] [CrossRef] [Green Version]
- Richardson, D.; Kalinowski, D.; Lau, S.; Jansson, P.; Lovejoy, D. Cancer cell iron metabolism and the development of potent iron chelators as anti-tumour agents. Biochim. Biophys. Acta 2009, 1790, 702–717. [Google Scholar] [CrossRef]
- Mertens, C.; Akam, E.A.; Rehwald, C.; Brüne, B.; Tomat, E.; Jung, M. Intracellular Iron Chelation Modulates the Macrophage Iron Phenotype with Consequences on Tumor Progression. PLoS ONE 2016, 11, e0166164. [Google Scholar] [CrossRef] [Green Version]
- Recalcati, S.; Locati, M.; Marini, A.; Santambrogio, P.; Zaninotto, F.; De Pizzol, M.; Zammataro, L.; Girelli, D.; Cairo, G. Differential regulation of iron homeostasis during human macrophage polarized activation. Eur. J. Immunol. 2010, 40, 824–835. [Google Scholar] [CrossRef]
- Pan, X.; Tamilselvam, B.; Hansen, E.J.; Daefler, S. Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 2010, 10, 64. [Google Scholar] [CrossRef] [Green Version]
- Harrington, N.P.; Chambers, K.A.; Ross, W.M.; Filion, L.G. Radiation damage and immune suppression in splenic mononuclear cell populations. Clin. Exp. Immunol. 1997, 107, 417–424. [Google Scholar] [CrossRef]
- Chen, Z.; Nomura, J.; Suzuki, T.; Suzuki, N. Enhanced Expression of Transferrin Receptor Confers UV-resistance in Human and Monkey Cells. J. Radiat. Res. 2005, 46, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Kenneth, N.S.; Mudie, S.; Naron, S.; Rocha, S. TfR1 interacts with the IKK complex and is involved in IKK–NF-κB signalling. Biochem. J. 2013, 449, 275–284. [Google Scholar] [CrossRef]






| UP Acession Number | Protein | Name | Gene | Average Ratio 1 | Std | Biological Process | Cellular Comp onent | Molecular Function |
|---|---|---|---|---|---|---|---|---|
| P62861 | RS30 | 40 S ribosomal protein S30 | FAU | 0.24 | 0.11 | Cytoplasmic translation, innate immune response in mucosa | Nucleus | Ribonucleoprotein |
| P29401 | TKT | Transketolase | TKT | 0.33 | 0.09 | Pentose-phosphate shunt | Nucleoplasm, peroxisome, endoplasmic reticulum membrane | Transferase |
| P67936 | TPM4 | Tropomyosin alpha-4 chain | TPM4 | 0.35 | 0.06 | Actin filament organization | Cytoskeleton | Actin-binding |
| P04080 | CYTB | Cystatin-B | CSTB | 0.35 | 0.24 | Negative regulation of proteolysis | Nucleus, Cytoplasm | Protease inhibitor |
| P07108 | ACBP | Acyl-CoA-binding protein | DBI | 0.36 | 0.38 | Fatty acid metabolic process | Endoplasmic reticulum, Golgi apparatus | Receptor |
| P02654 | APOC1 | Apolipoprotein C-I | APOC1 | 0.38 | 0.34 | Lipid transport | Secreted | Fatty acid binding |
| P07339 | CATD | Cathepsin D | CTSD | 0.40 | 0.27 | Autophagosome assembly, positive regulation of apoptotic process | Lysosome, Secreted | Aspartyl protease |
| P26038 | MOES | Moesin | MSN | 0.43 | 0.15 | Cytoskeleton organization | Cytoskeleton, Membrane | Actin binding |
| P11021 | GRP78 | 78 kDa glucose-regulated protein | HSPA5 | 0.52 | 0.09 | Cellular response to unfolded protein | Endoplasmic reticulum | Chaperone, Hydrolase |
| P04083 | ANXA1 | Annexin A1 | ANXA1 | 0.54 | 0.21 | Immunity, inflammatory response | Membrane, Nucleus, Cytoplasm, Secreted | Phospholipase A2 inhibitor |
| P63104 | 1433 Z | 14-3-3 protein zeta/delta | YWHAZ | 0.54 | 0.11 | Negative regulation of apoptosis, protein localization | Cytoplasm | Kinase, Monooxygenase |
| P19338 | NUCL | Nucleolin | NCL | 0.56 | 0.17 | Negative regulation of translation | Nucleus, Cytoplasm | DNA-binding, RNA-binding |
| O43175 | SERA | D-3-phosphoglycerate dehydrogenase | PHGDH | 0.59 | 0.29 | Amino-acid biosynthesis | Extracellular exosome | Oxidoreductase |
| P31948 | STIP1 | Stress-induced-phosphoprotein 1 | STIP1 | 0.59 | 0.16 | Protein folding | Nucleus, Cytoplasm | RNA-binding |
| Q09666 | AHNK | Neuroblast differentiation-associated protein AHNAK | AHNAK | 0.61 | 0.25 | Regulation of voltage-gated calcium channel activity | Nucleus, Virion | Viral nucleoprotein |
| Q15149 | PLEC | Plectin | PLEC | 0.62 | 0.11 | Cell morphogenesis | Cytoskeleton | Actin-binding |
| Q01469 | FABP5 | Fatty acid-binding protein, epidermal | FABP5 | 0.63 | 0.12 | Lipid transport | Nucleus, Cytoplasm, Secreted | Lipid binding |
| P21333 | FLNA | Filamin-A | FLNA | 0.63 | 0.24 | Actin cytoskeleton organization | Cytoskeleton | Actin-binding |
| P30456 | 1 A43 | HLA class I histocompatibility antigen, A-43 alpha chain | HLA-A | 0.64 | 0.30 | without GO annotation | ||
| Q5 JTZ9 | SYAM | Alanine—tRNA ligase, mitochondrial | AARS2 | 1.51 | 0.66 | Protein biosynthesis | Mitochondrion | Aminoacyl-tRNA synthetase |
| P05023 | AT1 A1 | Na+/K+-transporting ATPase subunit alpha-1 | ATP1 A1 | 1.80 | 0.09 | Ion transport | Membrane, Cell projection | Translocase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, A.T.; Machado, A.B.; Osório, H.; Pinto, M.L.; Vitorino, R.; Justino, G.; Santa, C.; Castro, F.; Cruz, T.; Rodrigues, C.; et al. Macrophage Resistance to Ionizing Radiation Exposure Is Accompanied by Decreased Cathepsin D and Increased Transferrin Receptor 1 Expression. Cancers 2023, 15, 270. https://doi.org/10.3390/cancers15010270
Pinto AT, Machado AB, Osório H, Pinto ML, Vitorino R, Justino G, Santa C, Castro F, Cruz T, Rodrigues C, et al. Macrophage Resistance to Ionizing Radiation Exposure Is Accompanied by Decreased Cathepsin D and Increased Transferrin Receptor 1 Expression. Cancers. 2023; 15(1):270. https://doi.org/10.3390/cancers15010270
Chicago/Turabian StylePinto, Ana Teresa, Ana Beatriz Machado, Hugo Osório, Marta Laranjeiro Pinto, Rui Vitorino, Gonçalo Justino, Cátia Santa, Flávia Castro, Tânia Cruz, Carla Rodrigues, and et al. 2023. "Macrophage Resistance to Ionizing Radiation Exposure Is Accompanied by Decreased Cathepsin D and Increased Transferrin Receptor 1 Expression" Cancers 15, no. 1: 270. https://doi.org/10.3390/cancers15010270