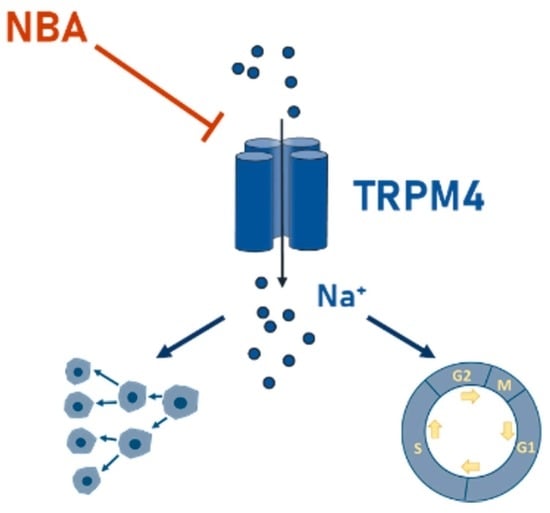
Investigation of Novel Small Molecular TRPM4 Inhibitors in Colorectal Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Electrophysiology
2.3. Drug Treatment
2.4. Viability Assay
2.5. Proliferation Assay
2.6. Cell Cycle Analysis
3. Results
3.1. Novel TRPM4 Inhibitors Block Endogenous TRPM4 Currents in CRC Cells

3.2. Novel TRPM4 Inhibitors Show No Effect on HCT116 Viability
3.3. TRPM4 Inhibitors Decrease the Proliferation of HCT116 Cells
3.4. NBA Affects Cell Cycle in HCT116 Cells
3.5. NBA Reduces the Viability of Colo205 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prevarskaya, N.; Ouadid-Ahidouch, H.; Skryma, R.; Shuba, Y. Remodelling of Ca2+ Transport in Cancer: How It Contributes to Cancer Hallmarks? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilius, B.; Prenen, J.; Janssens, A.; Owsianik, G.; Wang, C.; Zhu, M.X.; Voets, T. The Selectivity Filter of the Cation Channel TRPM4. J. Biol. Chem. 2005, 280, 22899–22906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Launay, P.; Fleig, A.; Perraud, A.L.; Scharenberg, A.M.; Penner, R.; Kinet, J.P. TRPM4 Is a Ca2+-Activated Nonselective Cation Channel Mediating Cell Membrane Depolarization. Cell 2002, 109, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Launay, P.; Cheng, H.; Srivatsan, S.; Penner, R.; Fleig, A.; Kinet, J.-P. TRPM4 Regulates Calcium Oscillations after T Cell Activation. Science 2004, 306, 1374–1377. [Google Scholar] [CrossRef] [Green Version]
- Fleig, A.; Penner, R. The TRPM Ion Channel Subfamily: Molecular, Biophysical and Functional Features. Trends Pharmacol. Sci. 2004, 25, 633–639. [Google Scholar] [CrossRef]
- Holzmann, C.; Kappel, S.; Kilch, T.; Jochum, M.M.; Urban, S.K.; Jung, V.; Stöckle, M.; Rother, K.; Greiner, M.; Peinelt, C. Transient Receptor Potential Melastatin 4 Channel Contributes to Migration of Androgen-Insensitive Prostate Cancer Cells. Oncotarget 2015, 6, 41783–41793. [Google Scholar] [CrossRef]
- Vennekens, R.; Olausson, J.; Meissner, M.; Bloch, W.; Mathar, I.; Philipp, S.E.; Schmitz, F.; Weissgerber, P.; Nilius, B.; Flockerzi, V.; et al. Increased IgE-Dependent Mast Cell Activation and Anaphylactic Responses in Mice Lacking the Calcium-Activated Nonselective Cation Channel TRPM4. Nat. Immunol. 2007, 8, 312–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Owsianik, G.; Freichel, M.; Flockerzi, V.; Nilius, B.; Vennekens, R. TRPM4 Regulates Migration of Mast Cells in Mice. Cell Calcium 2009, 45, 226–232. [Google Scholar] [CrossRef]
- Liu, H.; Chatel, S.; Simard, C.; Syam, N.; Salle, L.; Probst, V.; Morel, J.; Millat, G.; Lopez, M.; Abriel, H.; et al. Molecular Genetics and Functional Anomalies in a Series of 248 Brugada Cases with 11 Mutations in the TRPM4 Channel. PLoS ONE 2013, 8, e54131. [Google Scholar] [CrossRef] [Green Version]
- Kruse, M.; Schulze-Bahr, E.; Corfield, V.; Beckmann, A.; Stallmeyer, B.; Kurtbay, G.; Ohmert, I.; Schulze-Bahr, E.; Brink, P.; Pongs, O. Impaired Endocytosis of the Ion Channel TRPM4 Is Associated with Human Progressive Familial Heart Block Type I. J. Clin. Investig. 2009, 119, 2737–2744. [Google Scholar] [CrossRef]
- Kappel, S.; Stokłosa, P.; Hauert, B.; Ross-Kaschitza, D.; Borgström, A.; Baur, R.; Galván, J.A.; Zlobec, I.; Peinelt, C. TRPM4 Is Highly Expressed in Human Colorectal Tumor Buds and Contributes to Proliferation, Cell Cycle, and Invasion of Colorectal Cancer Cells. Mol. Oncol. 2019, 13, 2393–2405. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liao, P. TRPM4 Channel and Cancer. Cancer Lett. 2019, 454, 66–69. [Google Scholar] [CrossRef]
- Demion, M.; Bois, P.; Launay, P.; Guinamard, R. TRPM4, a Ca2+-Activated Nonselective Cation Channel in Mouse Sino-Atrial Node Cells. Cardiovasc. Res. 2007, 73, 531–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, K.D.; Soldini, D.; Jung, M.; Dietrich, D.; Stephan, C.; Jung, K.; Dietel, M.; Vainer, B.; Kristiansen, G. TRPM4 Protein Expression in Prostate Cancer: A Novel Tissue Biomarker Associated with Risk of Biochemical Recurrence Following Radical Prostatectomy. Virchows Arch. 2016, 468, 345–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbet, G.; Demion, M.; Moura, I.C.; Serafini, N.; Léger, T.; Vrtovsnik, F.; Monteiro, R.C.; Guinamard, R.; Kinet, J.-P.; Launay, P. The Calcium-Activated Nonselective Cation Channel TRPM4 Is Essential for the Migration but Not the Maturation of Dendritic Cells. Nat. Immunol. 2008, 9, 1148–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, M.; Pongs, O. TRPM4 Channels in the Cardiovascular System. Curr. Opin. Pharmacol. 2014, 15, 68–73. [Google Scholar] [CrossRef]
- Syam, N.; Chatel, S.; Ozhathil, L.C.; Sottas, V.; Rougier, J.-S.; Baruteau, A.; Baron, E.; Amarouch, M.-Y.; Daumy, X.; Probst, V.; et al. Variants of Transient Receptor Potential Melastatin Member 4 in Childhood Atrioventricular Block. J. Am. Heart Assoc. 2016, 5, e001625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; El Zein, L.; Kruse, M.; Guinamard, R.; Beckmann, A.; Bozio, A.; Kurtbay, G.; Mégarbané, A.; Ohmert, I.; Blaysat, G.; et al. Gain-of-Function Mutations in TRPM4 Cause Autosomal Dominant Isolated Cardiac Conduction Disease. Circ. Cardiovasc. Genet. 2010, 3, 374–385. [Google Scholar] [CrossRef] [Green Version]
- Ozhathil, L.C.; Delalande, C.; Bianchi, B.; Nemeth, G.; Kappel, S.; Thomet, U.; Ross-Kaschitza, D.; Simonin, C.; Rubin, M.; Gertsch, J.; et al. Identification of Potent and Selective Small Molecule Inhibitors of the Cation Channel TRPM4. Br. J. Pharmacol. 2018, 175, 2504–2519. [Google Scholar] [CrossRef] [Green Version]
- Schattling, B.; Steinbach, K.; Thies, E.; Kruse, M.; Menigoz, A.; Ufer, F.; Flockerzi, V.; Brück, W.; Pongs, O.; Vennekens, R.; et al. TRPM4 Cation Channel Mediates Axonal and Neuronal Degeneration in Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. Nat. Med. 2012, 18, 1805–1811. [Google Scholar] [CrossRef]
- Sagredo, A.I.; Sagredo, E.A.; Pola, V.; Echeverría, C.; Andaur, R.; Michea, L.; Stutzin, A.; Simon, F.; Marcelain, K.; Armisén, R. TRPM4 Channel Is Involved in Regulating Epithelial to Mesenchymal Transition, Migration, and Invasion of Prostate Cancer Cell Lines. J. Cell. Physiol. 2019, 234, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Sagredo, A.I.; Sagredo, E.A.; Cappelli, C.; Báez, P.; Andaur, R.E.; Blanco, C.; Tapia, J.C.; Echeverría, C.; Cerda, O.; Stutzin, A.; et al. TRPM4 Regulates Akt/GSK3-β Activity and Enhances β-Catenin Signaling and Cell Proliferation in Prostate Cancer Cells. Mol. Oncol. 2018, 12, 151–165. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Yu, J.-J. MicroRNA-150 Suppresses Epithelial-Mesenchymal Transition, Invasion, and Metastasis in Prostate Cancer through the TRPM4-Mediated β-Catenin Signaling Pathway. Am. J. Physiol. Physiol. 2018, 316, C463–C480. [Google Scholar] [CrossRef]
- Wong, K.K.; Hussain, F.A. TRPM4 Is Overexpressed in Breast Cancer Associated with Estrogen Response and Epithelial-Mesenchymal Transition Gene Sets. PLoS ONE 2020, 15, e0233884. [Google Scholar] [CrossRef]
- Loo, S.K.; Ch’ng, E.S.; Md Salleh, M.S.; Banham, A.H.; Pedersen, L.M.; Møller, M.B.; Green, T.M.; Wong, K.K. TRPM4 Expression Is Associated with Activated B Cell Subtype and Poor Survival in Diffuse Large B Cell Lymphoma. Histopathology 2017, 71, 98–111. [Google Scholar] [CrossRef]
- Armisén, R.; Marcelain, K.; Simon, F.; Tapia, J.C.; Toro, J.; Quest, A.F.G.; Stutzin, A. TRPM4 Enhances Cell Proliferation through Up-Regulation of the β-Catenin Signaling Pathway. J. Cell. Physiol. 2011, 226, 103–109. [Google Scholar] [CrossRef]
- Koelzer, V.H.; Zlobec, I.; Lugli, A. Tumor Budding in Colorectal Cancer—Ready for Diagnostic Practice? Hum. Pathol. 2016, 47, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Baker, K.; Minoo, P.; Hayashi, S.; Terracciano, L.; Lugli, A. Tumor Border Configuration Added to TNM Staging Better Stratifies Stage II Colorectal Cancer Patients into Prognostic Subgroups. Cancer 2009, 115, 4021–4029. [Google Scholar] [CrossRef]
- Guinamard, R.; Hof, T.; Del Negro, C.A. The TRPM4 Channel Inhibitor 9-Phenanthrol. Br. J. Pharmacol. 2014, 171, 1600–1613. [Google Scholar] [CrossRef] [Green Version]
- Grand, T.; Demion, M.; Norez, C.; Mettey, Y.; Launay, P.; Becq, F.; Bois, P.; Guinamard, R. 9-Phenanthrol Inhibits Human TRPM4 but Not TRPM5 Cationic Channels. Br. J. Pharmacol. 2008, 153, 1697–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garland, C.J.; Smirnov, S.V.; Bagher, P.; Lim, C.S.; Huang, C.Y.; Mitchell, R.; Stanley, C.; Pinkney, A.; Dora, K.A. TRPM4 Inhibitor 9-Phenanthrol Activates Endothelial Cell Intermediate Conductance Calcium-Activated Potassium Channels in Rat Isolated Mesenteric Artery. Br. J. Pharmacol. 2015, 172, 1114–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardam, K.E.; Geiger, J.E.; Hickey, C.M.; Hung, A.Y.; Magoski, N.S. Flufenamic Acid Affects Multiple Currents and Causes Intracellular Ca2+ Release in Aplysia Bag Cell Neurons. J. Neurophysiol. 2008, 100, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.G. KATP Channels as Molecular Sensors of Cellular Metabolism. Nature 2006, 440, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Borgström, A.; Hauert, B.; Kappel, S.; Zoni, E.; Kiener, M.; Stokłosa, P.; Baur, R.; Spahn, M.; Kruithof-de Julio, M.; Peinelt, C. Small Molecular Inhibitors Block TRPM4 Currents in Prostate Cancer Cells, with Limited Impact on Cancer Hallmark Functions. J. Mol. Biol. 2020, 433, 166665. [Google Scholar] [CrossRef]
- Rother, K.; Johne, C.; Spiesbach, K.; Haugwitz, U.; Tschöp, K.; Wasner, M.; Klein-Hitpass, L.; Möröy, T.; Mössner, J.; Engeland, K. Identification of Tcf-4 as a Transcriptional Target of P53 Signalling. Oncogene 2004, 23, 3376–3384. [Google Scholar] [CrossRef] [Green Version]
- Erwin Neher, Membranbiophysik MPI Göttingen, Copyright by F. M. and F. W. Patcher’s Power Tools Igor Pro XOPTM. Available online: https://www3.mpibpc.mpg.de/groups/neher/index.php?page=software (accessed on 1 June 2021).
- WEBMAXC STANDARD. Available online: https://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/webmaxc/webmaxcS.htm (accessed on 1 June 2021).
- Kho, D.; MacDonald, C.; Johnson, R.; Unsworth, C.P.; O’Carroll, S.J.; du Mez, E.; Angel, C.E.; Graham, E.S. Application of XCELLigence RTCA Biosensor Technology for Revealing the Profile and Window of Drug Responsiveness in Real Time. Biosensors 2015, 5, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Burris, S.K.; Wang, Q.; Bulley, S.; Neeb, Z.P.; Jaggar, J.H. 9-Phenanthrol Inhibits Recombinant and Arterial Myocyte TMEM16A Channels. Br. J. Pharmacol. 2015, 172, 2459–2468. [Google Scholar] [CrossRef] [Green Version]
- Veress, R.; Baranyai, D.; Hegyi, B.; Kistamás, K.; Dienes, C.; Magyar, J.; Bányász, T.; Nánási, P.P.; Szentandrássy, N.; Horváth, B. Transient Receptor Potential Melastatin 4 Channel Inhibitor 9-Phenanthrol Inhibits K(+) but Not Ca(2+) Currents in Canine Ventricular Myocytes. Can. J. Physiol. Pharmacol. 2018, 96, 1022–1029. [Google Scholar] [CrossRef]
- Low, S.W.; Gao, Y.; Wei, S.; Chen, B.; Nilius, B.; Liao, P. Development and Characterization of a Monoclonal Antibody Blocking Human TRPM4 Channel. Sci. Rep. 2021, 11, 10411. [Google Scholar] [CrossRef]
- Delalande, C.; Awale, M.; Rubin, M.; Probst, D.; Ozhathil, L.C.; Gertsch, J.; Abriel, H.; Reymond, J.-L. Optimizing TRPM4 Inhibitors in the MHFP6 Chemical Space. Eur. J. Med. Chem. 2019, 166, 167–177. [Google Scholar] [CrossRef]
- Gerzanich, V.; Woo, S.K.; Vennekens, R.; Tsymbalyuk, O.; Ivanova, S.; Ivanov, A.; Geng, Z.; Chen, Z.; Nilius, B.; Flockerzi, V.; et al. De Novo Expression of Trpm4 Initiates Secondary Hemorrhage in Spinal Cord Injury. Nat. Med. 2009, 15, 185–191. [Google Scholar] [CrossRef]
- Gorse, K.M.; Lantzy, M.K.; Lee, E.D.; Lafrenaye, A.D. Transient Receptor Potential Melastatin 4 Induces Astrocyte Swelling But Not Death after Diffuse Traumatic Brain Injury. J. Neurotrauma 2018, 35, 1694–1704. [Google Scholar] [CrossRef]
- Wang, C.; Naruse, K.; Takahashi, K. Role of the TRPM4 Channel in Cardiovascular Physiology and Pathophysiology. Cells 2018, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Gadd, S.; Narayan, G.; Bourdon, V.; Chaganti, S.; Arias-Pulido, H.; Nandula, S.V.; Rao, P.H.; Gissmann, L.; Dürst, M.; Schneider, A.; et al. Gene Dosage Alterations Revealed by CDNA Microarray Analysis in Cervical Cancer: Identification of Candidate Amplified and Overexpressed Genes. Genes Chromosom. Cancer 2007, 46, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Ceylan, G.G.; Önalan, E.E.; Kuloğlu, T.; Aydoğ, G.; Keleş, İ.; Tonyali, Ş.; Ceylan, C. Potential Role of Melastatin-Related Transient Receptor Potential Cation Channel Subfamily M Gene Expression in the Pathogenesis of Urinary Bladder Cancer. Oncol. Lett. 2016, 12, 5235–5239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borgström, A.; Peinelt, C.; Stokłosa, P. TRPM4 in Cancer-A New Potential Drug Target. Biomolecules 2021, 11, 229. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stokłosa, P.; Borgström, A.; Hauert, B.; Baur, R.; Peinelt, C. Investigation of Novel Small Molecular TRPM4 Inhibitors in Colorectal Cancer Cells. Cancers 2021, 13, 5400. https://doi.org/10.3390/cancers13215400
Stokłosa P, Borgström A, Hauert B, Baur R, Peinelt C. Investigation of Novel Small Molecular TRPM4 Inhibitors in Colorectal Cancer Cells. Cancers. 2021; 13(21):5400. https://doi.org/10.3390/cancers13215400
Chicago/Turabian StyleStokłosa, Paulina, Anna Borgström, Barbara Hauert, Roland Baur, and Christine Peinelt. 2021. "Investigation of Novel Small Molecular TRPM4 Inhibitors in Colorectal Cancer Cells" Cancers 13, no. 21: 5400. https://doi.org/10.3390/cancers13215400