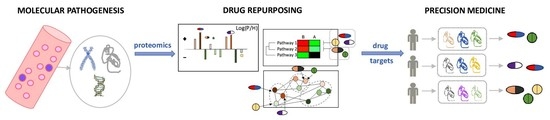
Proteomics and Drug Repurposing in CLL towards Precision Medicine
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Currently Known Pathophysiology, Molecular Diagnosis and Treatment Strategies in CLL
1.2. The Knowledge Gap in the Fight against CLL
1.3. Proteomics and Drug Repurposing in the Fight against CLL
2. Application of State-of-the-Art Mass Spectrometry-Based Proteomics in CLL Studies
2.1. The Powerful MS-Based Proteomics
2.2. Revelation of CLL through Proteomics
2.3. Proteomic Studies in CLL
2.3.1. Proteomic Studies Associated with IGHV Mutational Status
2.3.2. Proteomic Studies Associated with BCR Signaling
2.3.3. Proteomics Insights into Cytogenetics and Driver Mutations
2.3.4. Proteomic Approaches for Profiling Histones
2.3.5. Deciphering Cancerous Microenvironment Interactions through Proteomics
2.3.6. Subcellular Proteomics Studies
2.3.7. CLL Prognostic and Diagnostic Biomarkers by Proteomics Analysis
2.3.8. Specific Proteome Signature of CLL vs. Normal B Cells or Other Diseases
2.3.9. Proteomic Analysis after Pharmaceutical Treatment
2.3.10. Drug Repurposing Based on Proteomic Studies in CLL
2.3.11. Reverse-Phase Protein Array (RPPA) Studies in CLL
3. Drug Repurposing in CLL
3.1. Drug Repurposing in Hematological Malignancies—The Performance of CLL
3.2. Drug Repurposing Methodologies
3.2.1. In Silico—Computational Analysis
3.2.2. In Vitro—Experimental Analysis
3.2.3. Pros and Cons of Drug Repurposing Methodologies
3.2.4. The Proposed Pipeline
- (1)
- (Omic) data collection of the disease of interest (proteomics data are preferred).
- (2)
- Data classification and selection depending on the biological question.
- (3)
- Comparison of the data (e.g., with control/healthy donors or other diseases).
- (4)
- Functional and network analysis of the identified differences to unravel the deregulated pathways.
- (5)
- Identification of druggable targets and their association with FDA-approved drugs or drugs licensed for clinical trials (drug repurposing).
- (6)
- Experimental validation that the drugs have the expected properties in a simple disease model (cell culture).
- (7)
4. Proteomics-Based Drug Repurposing towards Precision Medicine in CLL
5. Conclusions
- Rearrange the already FDA-approved for CLL drugs to be given only in specific molecular traits, avoiding drug resistance.
- Align the drugs licensed for clinical trials in CLL to specific subtypes and stages.
- Find new drugs worthy of further study for the treatment of CLL.
- Make more effective drug combinations that can either target the most deregulated pathways or may have a synergism on a specific pathway/active subnetwork/co-expression network or even show a balancing action on toxicity effects/drug resistance, based both on the molecular signature of the disease and on the networks induced by the administration of the drugs.
6. Future Perspectives and Challenges
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MS/MS | tandem mass spectrometry |
| PTM | post translation modifications |
| M | mutated |
| U | unmutated |
| P | patient |
| progres. | progressive |
| indol. | indolent |
| SLL | small lymphocytic lymphoma |
| ALL | acute lymphoblastic leukemia |
| AML | acute myeloid leukemia |
| BL | Burkitt lymphoma |
| CML | chronic myeloid leukemia |
| DLBCL | diffuse large B cell lymphoma |
| HD | healthy donors |
| RPPA | Reverse Phase Protein Array |
| Cys | Cysteine |
References
- Mosquera Orgueira, A.; Antelo Rodríguez, B.; Díaz Arias, J.; González Pérez, M.S.; Bello López, J.L. New Recurrent Structural Aberrations in the Genome of Chronic Lymphocytic Leukemia Based on Exome-Sequencing Data. Front. Genet. 2019, 10, 854. [Google Scholar] [CrossRef] [Green Version]
- Rai, K.R.; Jain, P. Chronic lymphocytic leukemia (CLL)-Then and now. Am. J. Hematol. 2016, 91, 330–340. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Kipps, T.J. The pathogenesis of chronic lymphocytic leukemia. Annu. Rev. Pathol. 2014, 9, 103–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brieghel, C.; da Cunha-Bang, C.; Yde, C.W.; Schmidt, A.Y.; Kinalis, S. The Number of Signaling Pathways Altered by Driver Mutations in Chronic Lymphocytic Leukemia Impacts Disease Outcome. Clin. Cancer Res. 2020, 26, 1507–1515. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.C.; Margolskee, E. Chronic lymphocytic leukemia with TP53 gene alterations: A detailed clinicopathologic analysis. Mod. Pathol. 2020, 33, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Malcikova, J.; Tausch, E.; Rossi, D.; Sutton, L.A.; Soussi, T. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia 2018, 32, 1070–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelmann, J.; Holzmann, K.; Tausch, E.; Saunderson, E.A.; Jebaraj, B.M.C.; Steinbrecher, D.; Dolnik, A.; Blätte, T.J.; Landau, D.A.; Saub, J.; et al. Genomic alterations in high-risk chronic lymphocytic leukemia frequently affect cell cycle key regulators and NOTCH1-regulated transcription. Haematologica 2020, 105, 1379–1390. [Google Scholar] [CrossRef]
- Chiorazzi, N.; Rai, K.R.; Ferrarini, M. Chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 352, 804–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, L.D.; Minton, A.R.; Blunt, M.D.; Karydis, L.I.; Dutton, D.A.; Rogers-Broadway, K.R.; Dobson, R.; Liu, R.; Norster, F.; Hogg, E. BCR signaling contributes to autophagy regulation in chronic lymphocytic leukemia. Leukemia 2020, 34, 640–644. [Google Scholar] [CrossRef]
- Rigolin, G.M.; Saccenti, E.; Guardalben, E.; Cavallari, M.; Formigaro, L.; Zagatti, B.; Visentin, A.; Mauro, F.R. In chronic lymphocytic leukaemia with complex karyotype, major structural abnormalities identify a subset of patients with inferior outcome and distinct biological characteristics. Br. J. Haematol. 2018, 181, 229–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baliakas, P.; Jeromin, S.; Iskas, M.; Puiggros, A.; Plevova, K.; Nguyen-Khac, F.; Davis, Z.; Rigolin, G.M.; Visentin, A.; Xochelli, A.; et al. Cytogenetic complexity in chronic lymphocytic leukemia: Definitions, associations, and clinical impact. Blood 2019, 133, 1205–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visentin, A.; Bonaldi, L.; Rigolin, G.M.; Mauro, F.R.; Martines, A.; Frezzato, F.; Imbergamo, S.; Scomazzon, E.; Pravato, S.; Bardi, M.A.; et al. The combination of complex karyotype subtypes and IGHV mutational status identifies new prognostic and predictive groups in chronic lymphocytic leukaemia. Br. J. Cancer 2019, 121, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Leeksma, A.C.; Baliakas, P.; Moysiadis, T.; Puiggros, A.; Plevova, K.; Van der Kevie-Kersemaekers, A.M.; Posthuma, H.; Rodriguez-Vicente, A.E.; Tran, A.N.; Barbany, G.; et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: A multi-center study. Haematologica 2021, 106, 87–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jondreville, L.; Krzisch, D.; Chapiro, E. The complex karyotype and chronic lymphocytic leukemia: Prognostic value and diagnostic recommendations. Am. J. Hematol. 2020, 95, 1361–1367. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, S.; Banerji, V. Targeting Mitochondrial Bioenergetics as a Therapeutic Strategy for Chronic Lymphocytic Leukemia. Oxid. Med. Cell. Longev. 2018, 2426712. [Google Scholar] [CrossRef]
- Van Attekum, M.H.; Eldering, E.; Kater, A.P. Chronic lymphocytic leukemia cells are active participants in microenvironmental cross-talk. Haematologica 2017, 102, 1469–1476. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Trachootham, D.; Liu, J.; Chen, G.; Pelicano, H.; Garcia-Prieto, C.; Lu, W.; Burger, J.A.; Croce, C.M.; Plunkett, W.; et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat. Cell Biol. 2012, 14, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Jitschin, R.; Braun, M.; Qorraj, M.; Saul, D.; Le Blanc, K.; Zenz, T.; Mougiakakos, D. Stromal cell-mediated glycolytic switch in CLL cells involves Notch-c-Myc signaling. Blood 2015, 125, 3432–3436. [Google Scholar] [CrossRef] [Green Version]
- Sharif-Askari, B.; Doyon, D.; Paliouras, M. Bruton’s tyrosine kinase is at the crossroads of metabolic adaptation in primary malignant human lymphocytes. Sci. Rep. 2019, 9, 11069. [Google Scholar] [CrossRef] [Green Version]
- Binet, J.L.; Auquier, A.; Dighiero, G.; Chastang, C.; Piguet, H.; Goasguen, J.; Vaugier, G.; Potron, G.; Colona, P.; Oberling, F.; et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 1981, 48, 198–206. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef] [Green Version]
- PDQ Supportive and Palliative Care Editorial Board. Chronic Lymphocytic Leukemia Treatment (PDQ®): Health Professional Version. In PDQ Cancer Information Summaries; National Cancer Institute (US): Bethesda, MD, USA, 2002. [Google Scholar]
- Falay, M.; Serdar, M.A.; Dalgali, H.; Uçar, M.A.; Dagdaş, S.; Özet, G. Which Markers Should the used for Diagnostic Chronic Lymphocytic Leukemia Immunophenotyping Scoring System by Flow Cytometry? Clin. Lab. 2019, 65. [Google Scholar] [CrossRef]
- Sorigue, M.; Magnano, L.; Miljkovic, M.D.; Nieto-Moragas, J.; Santos-Gomez, M.; Villamor, N.; Junca, J.; Morales-Indiano, C. Positive predictive value of CD200 positivity in the differential diagnosis of chronic lymphocytic leukemia. Cytom. B Clin. Cytom. 2020, 98, 441–448. [Google Scholar] [CrossRef]
- Myles, N.; Giri, P.; Chim, I.; Kodituwakku, A. The utility of CD200 expression and modified Matutes score in the diagnostic differentiation of mantle cell lymphoma and chronic lymphocytic leukemia using flow cytometry. Leuk. Lymphoma 2021, 62, 504–506. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Huang, X.; Ye, X.; Qian, W. Prognostic and clinicopathological significance of PD-1/PD-L1 expression in the tumor microenvironment and neoplastic cells for lymphoma. Int. Immunopharmacol. 2019, 77, 105999. [Google Scholar] [CrossRef] [PubMed]
- Mosquera Orgueira, A.; Antelo Rodríguez, B.; Díaz Arias, J.; Bello López, J.L. Identification of new putative driver mutations and predictors of disease evolution in chronic lymphocytic leukemia. Blood Cancer J. 2019, 9, 78. [Google Scholar] [CrossRef] [Green Version]
- Katsaraki, K.; Artemaki, P.I.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. Identification of a novel, internal tRNA-derived RNA fragment as a new prognostic and screening biomarker in chronic lymphocytic leukemia, using an innovative quantitative real-time PCR assay. Leuk. Res. 2019, 87, 106234. [Google Scholar] [CrossRef] [PubMed]
- Loi, E.; Moi, L.; Fadda, A.; Satta, G.; Zucca, M.; Sanna, S.; Amini Nia, S.; Cabras, G.; Padoan, M.; Magnani, C.; et al. Methylation alteration of SHANK1 as a predictive, diagnostic and prognostic biomarker for chronic lymphocytic leukemia. Oncotarget 2019, 10, 4987–5002. [Google Scholar] [CrossRef] [Green Version]
- Casabonne, D.; Benavente, Y.; Seifert, J.; Costas, L.; Armesto, M.; Arestin, M.; Besson, C. Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int. J. Cancer 2020, 147, 1315–1324. [Google Scholar] [CrossRef]
- Del Giudice, I.; Raponi, S.; Della Starza, I.; De Propris, M.S.; Cavalli, M.; De Novi, L.A.; Cappelli, L.V.; Ilari, C.; Cafforio, L.; Guarini, A.; et al. Minimal Residual Disease in Chronic Lymphocytic Leukemia: A New Goal? Front. Oncol. 2019, 9, 689. [Google Scholar] [CrossRef] [Green Version]
- Jain, N.; O’Brien, S. Targeted therapies for CLL: Practical issues with the changing treatment paradigm. Blood Rev. 2016, 30, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.L.; Gribben, J.G. Immunotherapy in Chronic Lymphocytic Leukaemia (CLL). Curr. Hematol. Malig. Rep. 2016, 11, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.H.; Niesen, E.; Hallek, M. New roles for B cell receptor associated kinases: When the B cell is not the target. Leukemia 2019, 33, 576–587. [Google Scholar] [CrossRef]
- Schiattone, L.; Ghia, P.; Scarfò, L. The evolving treatment landscape of chronic lymphocytic leukemia. Curr. Opin. Oncol. 2019, 31, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Maldonado, V.; García-Morillo, M.; Delgado, J. The biology behind PI3K inhibition in chronic lymphocytic leukaemia. Ther. Adv. Hematol. 2015, 6, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perini, G.F.; Ribeiro, G.N.; Pinto Neto, J.V.; Campos, L.T.; Hamerschlak, N. BCL-2 as therapeutic target for hematological malignancies. J. Hematol. Oncol. 2018, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Sharman, J.; Di Paolo, J. Targeting B-cell receptor signaling kinases in chronic lymphocytic leukemia: The promise of entospletinib. Ther. Adv. Hematol. 2016, 7, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bair, S.M.; Porter, D.L. Accelerating chimeric antigen receptor therapy in chronic lymphocytic leukemia: The development and challenges of chimeric antigen receptor T-cell therapy for chronic lymphocytic leukemia. Am. J. Hematol. 2019, 94, S10–S17. [Google Scholar] [CrossRef] [Green Version]
- Lemal, R.; Tournilhac, O. State-of-the-art for CAR T-cell therapy for chronic lymphocytic leukemia in 2019. J. Immunother. Ther. Cancer 2019, 7, 202. [Google Scholar] [CrossRef] [Green Version]
- Forte, D.; Krause, D.S.; Andreeff, M.; Bonnet, D.; Méndez-Ferrer, S. Updates on the hematologic tumor microenvironment and its therapeutic targeting. Haematologica 2019, 104, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulos, C.; Kostareli, E. Advances in Epigenetics and Epigenomics in Chronic Lymphocytic Leukemia. Curr. Genet. Med. Rep. 2019, 7, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Janovská, P.; Bryja, V. Wnt signalling pathways in chronic lymphocytic leukaemia and B-cell lymphomas. Br. J. Pharmacol. 2017, 174, 4701–4715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, M.; Sharma, A.R. Interaction between miRNAs and signaling cascades of Wnt pathway in chronic lymphocytic leukemia. J. Cell. Biochem. 2020, 121, 4654–4666. [Google Scholar] [CrossRef]
- Rosati, E.; Baldoni, S.; De Falco, F.; Del Papa, B.; Dorillo, E.; Rompietti, C.; Albi, E.; Falzetti, F.; Di Ianni, M.; Sportoletti, P. NOTCH1 Aberrations in Chronic Lymphocytic Leukemia. Front. Oncol. 2018, 8, 229. [Google Scholar] [CrossRef] [Green Version]
- Mansouri, L.; Wierzbinska, J.A.; Plass, C.; Rosenquist, R. Epigenetic deregulation in chronic lymphocytic leukemia: Clinical and biological impact. Semin. Cancer Biol. 2018, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Li-Talley, M.; Buck, A.; Chen, W. An emerging trend of rapid increase of leukemia but not all cancers in the aging population in the United States. Sci. Rep. 2019, 9, 12070. [Google Scholar] [CrossRef] [Green Version]
- Hallek, M. Chronic lymphocytic leukemia: 2017 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2017, 92, 946–965. [Google Scholar] [CrossRef]
- Mulligan, S.P.; Shumack, S.; Guminski, A. Chronic lymphocytic leukemia, skin and other second cancers. Leuk. Lymphoma 2019, 60, 3104–3106. [Google Scholar] [CrossRef]
- Kumar, V.; Ailawadhi, S.; Bojanini, L.; Mehta, A.; Biswas, S.; Sher, T.; Roy, V.; Vishnu, P.; Marin-Acevedo, J. Trends in the risk of second primary malignancies among survivors of chronic lymphocytic leukemia. Blood Cancer J. 2019, 9, 75. [Google Scholar] [CrossRef]
- Bond, D.A.; Huang, Y.; Fisher, J.L.; Ruppert, A.S.; Owen, D.H.; Bertino, E.M.; Rogers, K.A.; Bhat, S.A.; Grever, M.R.; Jaglowski, S.M.; et al. Second cancer incidence in CLL patients receiving BTK inhibitors. Leukemia 2020, 34, 3197–3205. [Google Scholar] [CrossRef] [PubMed]
- Van Der Nest, B.M.; Leslie, C.; Joske, D.; Radeski, D.; White, R.; Cheah, C.Y. Peripheral T-Cell Lymphoma Arising in Patients With Chronic Lymphocytic Leukemia. Am. J. Clin. Pathol. 2019, 152, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Enya Chen, Y.C.; Burgess, M.; Mapp, S.; Mollee, P.; Gill, D.; Blumenthal, A.; Saunders, N.A. PI3K-p110δ contributes to antibody responses by macrophages in chronic lymphocytic leukemia. Leukemia 2020, 34, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Alsagaby, S.A.; Alhumaydhi, F.A. Proteomics insights into the pathology and prognosis of chronic lymphocytic leukemia. Saudi Med. J. 2019, 40, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.E.; Carter, M.J.; Larrayoz, M.; Clarke, J.; Garbis, S.D.; Oscier, D.; Strefford, J.C.; Steele, A.J.; Walewska, R.; Cragg, M.S. Proteomics Profiling of CLL Versus Healthy B-cells Identifies Putative Therapeutic Targets and a Subtype-independent Signature of Spliceosome Dysregulation. Mol. Cell. Proteom. 2018, 17, 776–791. [Google Scholar] [CrossRef] [Green Version]
- Thurgood, L.A.; Chataway, T.K.; Lower, K.M.; Kuss, B.J. From genome to proteome: Looking beyond DNA and RNA in chronic lymphocytic leukemia. J. Proteom. 2017, 155, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Psatha, K.; Kollipara, L.; Voutyraki, C.; Divanach, P.; Sickmann, A.; Rassidakis, G.Z.; Drakos, E.; Aivaliotis, M. Deciphering lymphoma pathogenesis via state-of-the-art mass spectrometry-based quantitative proteomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1047, 2–14. [Google Scholar] [CrossRef]
- Gupta, A.; Kumar, A. Pros and cons of the proteomics. Biomed. J. 2014, 37, 163–164. [Google Scholar] [CrossRef]
- Díez, P.; Góngora, R.; Orfao, A.; Fuentes, M. Functional proteomic insights in B-cell chronic lymphocytic leukemia. Expert Rev. Proteom. 2017, 14, 137–146. [Google Scholar] [CrossRef]
- Almaiman, A.A. Proteomic Profile of Lymphoid Leukemia. J. Coll. Phys. Surg. Pak. 2018, 28, 133–145. [Google Scholar] [CrossRef]
- Cochran, D.A.; Evans, C.A.; Blinco, D.; Burthem, J.; Stevenson, F.K.; Gaskell, S.J.; Whetton, A.D. Proteomic analysis of chronic lymphocytic leukemia subtypes with mutated or unmutated Ig V(H) genes. Mol. Cell. Proteom. 2003, 2, 1331–1341. [Google Scholar] [CrossRef] [Green Version]
- Barnidge, D.R.; Jelinek, D.F.; Muddiman, D.C.; Kay, N.E. Quantitative protein expression analysis of CLL B cells from mutated and unmutated IgV(H) subgroups using acid-cleavable isotope-coded affinity tag reagents. J. Proteom. Res. 2005, 4, 1310–1317. [Google Scholar] [CrossRef] [Green Version]
- Rees-Unwin, K.S.; Faragher, R.; Unwin, R.D.; Adams, J.; Brown, P.J.; Buckle, A.M.; Pettitt, A.; Hutchinson, C.V.; Johnson, S.M.; Pulford, K.; et al. Ribosome-associated nucleophosmin 1: Increased expression and shuttling activity distinguishes prognostic subtypes in chronic lymphocytic leukaemia. Br. J. Haematol. 2010, 148, 534–543. [Google Scholar] [CrossRef]
- Eagle, G.L.; Zhuang, J.; Jenkins, R.E.; Till, K.J.; Jithesh, P.V.; Lin, K.; Johnson, G.G.; Oates, M.; Park, K.; Kitteringham, N.R.; et al. Total proteome analysis identifies migration defects as a major pathogenetic factor in immunoglobulin heavy chain variable region (IGHV)-unmutated chronic lymphocytic leukemia. Mol. Cell. Proteom. 2015, 14, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thurgood, L.A.; Dwyer, E.S.; Lower, K.M.; Chataway, T.K.; Kuss, B.J. Altered expression of metabolic pathways in CLL detected by unlabelled quantitative mass spectrometry analysis. Br. J. Haematol. 2019, 185, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Eagle, G.L.; Herbert, J.M.J.; Zhuang, J.; Oates, M.; Khan, U.T.; Kitteringham, N.R.; Clarke, K.; Park, B.K.; Pettitt, A.R.; Jenkins, R.E.; et al. Assessing technical and biological variation in SWATH-MS-based proteomic analysis of chronic lymphocytic leukaemia cells. Sci. Rep. 2021, 11, 2932. [Google Scholar] [CrossRef]
- Scielzo, C.; Ghia, P.; Conti, A.; Bachi, A.; Guida, G.; Geuna, M.; Alessio, M.; Caligaris-Cappio, F. HS1 protein is differentially expressed in chronic lymphocytic leukemia patient subsets with good or poor prognoses. J. Clin. Investig. 2005, 115, 1644–1650. [Google Scholar] [CrossRef]
- Perrot, A.; Pionneau, C.; Nadaud, S.; Davi, F.; Leblond, V.; Jacob, F.; Merle-Béral, H.; Herbrecht, R.; Béné, M.C.; Gribben, J.G.; et al. A unique proteomic profile on surface IgM ligation in unmutated chronic lymphocytic leukemia. Blood 2011, 118, e1–e15. [Google Scholar] [CrossRef] [Green Version]
- Alsagaby, S.A.; Khanna, S.; Hart, K.W.; Pratt, G.; Fegan, C.; Pepper, C.; Brewis, I.A.; Brennan, P. Proteomics-based strategies to identify proteins relevant to chronic lymphocytic leukemia. J. Proteom. Res. 2014, 13, 5051–5062. [Google Scholar] [CrossRef] [PubMed]
- Glibert, P.; Vossaert, L.; Van Steendam, K.; Lambrecht, S.; Van Nieuwerburgh, F.; Offner, F.; Kipps, T.; Dhaenens, M.; Deforce, D. Quantitative proteomics to characterize specific histone H2A proteolysis in chronic lymphocytic leukemia and the myeloid THP-1 cell line. Int. J. Mol. Sci. 2014, 15, 9407–9421. [Google Scholar] [CrossRef] [Green Version]
- Díez, P.; Lorenzo, S.; Dégano, R.M.; Ibarrola, N.; González-González, M.; Nieto, W.; Almeida, J.; González, M.; Orfao, A.; Fuentes, M. Multipronged functional proteomics approaches for global identification of altered cell signalling pathways in B-cell chronic lymphocytic leukaemia. Proteomics 2016, 16, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Kashuba, E.; Eagle, G.L.; Bailey, J.; Evans, P.; Welham, K.J.; Allsup, D.; Cawkwell, L. Proteomic analysis of B-cell receptor signaling in chronic lymphocytic leukaemia reveals a possible role for kininogen. J. Proteom. 2013, 91, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Díez, P.; Ibarrola, N.; Dégano, R.M.; Lécrevisse, Q.; Rodriguez-Caballero, A.; Criado, I.; Nieto, W.G.; Góngora, R.; González, M.; Almeida, J.; et al. A systematic approach for peptide characterization of B-cell receptor in chronic lymphocytic leukemia cells. Oncotarget 2017, 8, 42836–42846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, T.; Ahorn, H.; Haberl, P.; Döhner, H.; Wilgenbus, K. Correlation of clinical data with proteomics profiles in 24 patients with B-cell chronic lymphocytic leukemia. Int. J. Cancer 2001, 91, 180–186. [Google Scholar] [CrossRef]
- Huang, P.Y.; Mactier, S.; Armacki, N.; Giles Best, O.; Belov, L.; Kaufman, K.L.; Pascovici, D.; Mulligan, S.P.; Christopherson, R.I. Protein profiles distinguish stable and progressive chronic lymphocytic leukemia. Leuk. Lymphoma 2016, 57, 1033–1043. [Google Scholar] [CrossRef]
- Bretones, G.; Álvarez, M.G.; Arango, J.R.; Rodríguez, D.; Nadeu, F.; Prado, M.A.; Valdés-Mas, R.; Puente, D.A.; Paulo, J.A.; Delgado, J.; et al. Altered patterns of global protein synthesis and translational fidelity in RPS15-mutated chronic lymphocytic leukemia. Blood 2018, 132, 2375–2388. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Lucas, D.M.; Zhang, L.; Xu, H.; Zabrouskov, V.; Davis, M.E.; Knapp, A.R.; Young, D.C.; Payne, P.R.; Parthun, M.R.; et al. Validation of an LC-MS based approach for profiling histones in chronic lymphocytic leukemia. Proteomics 2009, 9, 1197–1206. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Harshman, S.W.; Ruppert, A.S.; Mortazavi, A.; Lucas, D.M.; Thomas-Ahner, J.M.; Clinton, S.K.; Byrd, J.C.; Freitas, M.A.; Parthun, M.R. Proteomic profiling identifies specific histone species associated with leukemic and cancer cells. Clin. Proteom. 2015, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- O’Hayre, M.; Salanga, C.L.; Kipps, T.J.; Messmer, D.; Dorrestein, P.C.; Handel, T.M. Elucidating the CXCL12/CXCR4 signaling network in chronic lymphocytic leukemia through phosphoproteomics analysis. PLoS ONE 2010, 5, e11716. [Google Scholar] [CrossRef] [Green Version]
- Prieto, D.; Sotelo, N.; Seija, N.; Sernbo, S.; Abreu, C.; Durán, R.; Gil, M.; Sicco, E.; Irigoin, V.; Oliver, C.; et al. S100-A9 protein in exosomes from chronic lymphocytic leukemia cells promotes NF-κB activity during disease progression. Blood 2017, 130, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Haderk, F.; Schulz, R. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangolini, M.; Götte, F.; Moore, A. Notch2 controls non-autonomous Wnt-signalling in chronic lymphocytic leukaemia. Nat. Commun. 2018, 9, 3839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, R.S.; Adam, P.J.; Patel, S.; Loader, J.A.; Berry, J.; Redpath, N.T.; Poyser, H.R.; Fletcher, G.C.; Burgess, N.A.; Stamps, A.C.; et al. Proteomic analysis of the cell-surface membrane in chronic lymphocytic leukemia: Identification of two novel proteins, BCNP1 and MIG2B. Leukemia 2003, 17, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miguet, L.; Béchade, G.; Fornecker, L.; Zink, E.; Felden, C.; Gervais, C.; Herbrecht, R.; Van Dorsselaer, A.; Mauvieux, L.; Sanglier-Cianferani, S. Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J. Proteom. Res. 2009, 8, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Henrich, S.; Crossett, B.; Christopherson, R.I. Differentially expressed nuclear proteins in human CCRF-CEM, HL-60, MEC-1 and Raji cells correlate with cellular properties. Proteom. Clin. Appl. 2007, 1, 1252–1265. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.L.; Schwarzmeier, J.D.; Gerner, M.C.; Bileck, A.; Mader, J.C.; Meier-Menches, S.M.; Gerner, S.M.; Schmetterer, K.G.; Pukrop, T.; Reichle, A.; et al. Proteomics and metabolomics identify molecular mechanisms of aging potentially predisposing for chronic lymphocytic leukemia. Mol. Cell. Proteom. 2018, 17, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Gez, S.; Crossett, B.; Christopherson, R.I. Differentially expressed cytosolic proteins in human leukemia and lymphoma cell lines correlate with lineages and functions. Biochim. Biophys. Acta 2007, 1774, 1173–1183. [Google Scholar] [CrossRef]
- Miguet, L.; Bogumil, R.; Decloquement, P.; Herbrecht, R.; Potier, N.; Mauvieux, L.; Van Dorsselaer, A. Discovery and identification of potential biomarkers in a prospective study of chronic lymphoid malignancies using SELDI-TOF-MS. J. Proteom. Res. 2006, 5, 2258–2269. [Google Scholar] [CrossRef]
- Schröder, C.; Srinivasan, H.; Sill, M.; Linseisen, J.; Fellenberg, K.; Becker, N.; Nieters, A.; Hoheisel, J.D. Plasma protein analysis of patients with different B-cell lymphomas using high-content antibody microarrays. Proteom. Clin. Appl. 2013, 7, 802–812. [Google Scholar] [CrossRef]
- Johnston, H.E.; Carter, M.J.; Cox, K.L.; Dunscombe, M.; Manousopoulou, A.; Townsend, P.A.; Garbis, S.D. Integrated Cellular and Plasma Proteomics of Contrasting B-cell Cancers Reveals Common, Unique and Systemic Signatures. Mol. Cell. Proteom. 2017, 16, 386–406. [Google Scholar] [CrossRef] [Green Version]
- Marina, O.; Biernacki, M.A.; Brusic, V.; Wu, C.J. A concentration-dependent analysis method for high density protein microarrays. J. Proteom. Res. 2008, 7, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Khodadoust, M.S.; Olsson, N. B-cell lymphomas present immunoglobulin neoantigens. Blood 2019, 133, 878–881. [Google Scholar] [CrossRef]
- Henrich, S.; Mactier, S.; Best, G.; Mulligan, S.P.; Crossett, B.; Christopherson, R.I. Fludarabine nucleoside modulates nuclear “survival and death” proteins in resistant chronic lymphocytic leukemia cells. Nucleosides Nucleotides Nucleic Acids 2011, 30, 1181–1189. [Google Scholar] [CrossRef]
- Che, Y.; Best, O.G.; Zhong, L.; Kaufman, K.L.; Mactier, S.; Raftery, M.; Graves, L.M.; Mulligan, S.P.; Christopherson, R.I. Hsp90 Inhibitor SNX-7081 dysregulates proteins involved with DNA repair and replication and the cell cycle in human chronic lymphocytic leukemia (CLL) cells. J. Proteom. Res. 2013, 12, 1710–1722. [Google Scholar] [CrossRef]
- Kaufman, K.L.; Jenkins, Y.; Alomari, M.; Mirzaei, M.; Best, O.G.; Pascovici, D.; Mactier, S.; Mulligan, S.P.; Haynes, P.A.; Christopherson, R.I. The Hsp90 inhibitor SNX-7081 is synergistic with fludarabine nucleoside via DNA damage and repair mechanisms in human, p53-negative chronic lymphocytic leukemia. Oncotarget 2015, 6, 40981–40997. [Google Scholar] [CrossRef]
- Kruse, U.; Pallasch, C.P.; Bantscheff, M.; Eberhard, D.; Frenzel, L.; Ghidelli, S.; Maier, S.K.; Werner, T.; Wendtner, C.M.; Drewes, G. Chemoproteomics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia 2011, 25, 89–100. [Google Scholar] [CrossRef]
- Beckmann, L.; Berg, V.; Dickhut, C.; Sun, C.; Merkel, O.; Bloehdorn, J.; Robrecht, S.; Seifert, M.; da Palma Guerreiro, A.; Claasen, J.; et al. MARCKS affects cell motility and response to BTK inhibitors in CLL. Blood 2021. [Google Scholar] [CrossRef]
- Shull, A.Y.; Noonepalle, S.K.; Awan, F.T.; Liu, J.; Pei, L.; Bollag, R.J.; Salman, H.; Ding, Z.; Shi, H. RPPA-based protein profiling reveals eIF4G overexpression and 4E-BP1 serine 65 phosphorylation as molecular events that correspond with a pro-survival phenotype in chronic lymphocytic leukemia. Oncotarget 2015, 6, 14632–14645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frezzato, F.; Accordi, B.; Trimarco, V.; Gattazzo, C.; Martini, V.; Milani, G.; Bresolin, S.; Severin, F.; Visentin, A.; Basso, G.; et al. Profiling B cell chronic lymphocytic leukemia by reverse phase protein array: Focus on apoptotic proteins. J. Leukoc. Biol. 2016, 100, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.K.; Lamothe, B.; Ayres, M.L.; Gay, J.; Cheung, J.P.; Balakrishnan, K.; Ivan, C. Pharmacodynamics and proteomic analysis of acalabrutinib therapy: Similarity of on-target effects to ibrutinib and rationale for combination therapy. Leukemia 2018, 32, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Vangapandu, H.V.; Havranek, O.; Ayres, M.L.; Kaipparettu, B.A.; Balakrishnan, K.; Wierda, W.G.; Keating, M.J.; Davis, R.E.; Stellrecht, C.M.; Gandhi, V. B-cell Receptor Signaling Regulates Metabolism in Chronic Lymphocytic Leukemia. Mol. Cancer Res. 2017, 15, 1692–1703. [Google Scholar] [CrossRef] [Green Version]
- Langedijk, J.; Mantel-Teeuwisse, A.K.; Slijkerman, D.S.; Schutjens, M.H. Drug repositioning and repurposing: Terminology and definitions in literature. Drug Discov. Today 2015, 20, 1027–1034. [Google Scholar] [CrossRef]
- Konc, J. Binding site comparisons for target-centered drug discovery. Expert Opin. Drug Discov. 2019, 14, 445–454. [Google Scholar] [CrossRef]
- McCabe, B.; Liberante, F.; Mills, K.I. Repurposing medicinal compounds for blood cancer treatment. Ann. Hematol. 2015, 94, 1267–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol. Sci. 2018, 14, 1232–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleire, L.; Førde, H.E.; Netland, I.A.; Leiss, L.; Skeie, B.S.; Enger, P. Drug repurposing in cancer. Pharmacol. Res. 2017, 124, 74–91. [Google Scholar] [CrossRef]
- Kaushik, I.; Ramachandran, S.; Prasad, S.; Srivastava, S.K. Drug rechanneling: A novel paradigm for cancer treatment. Semin. Cancer Biol. 2021, 68, 279–290. [Google Scholar] [CrossRef]
- Kirtonia, A.; Gala, K.; Fernandes, S.G.; Pandya, G.; Pandey, A.K.; Sethi, G.; Khattar, E.; Garg, M. Repurposing of drugs: An attractive pharmacological strategy for cancer therapeutics. Semin. Cancer Biol. 2021, 68, 258–278. [Google Scholar] [CrossRef]
- Orecchioni, S.; Roma, S.; Raimondi, S.; Gandini, S.; Bertolini, F. Identifying Drug Repurposing Opportunities in Oncology. Cancer J. 2019, 25, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Armando, R.G.; Mengual Gómez, D.L.; Gomez, D.E. New drugs are not enough-drug repositioning in oncology: An update. Int. J. Oncol. 2020, 56, 651–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olgen, S.; Kotra, L.P. Drug Repurposing in the Development of Anticancer Agents. Curr. Med. Chem. 2019, 26, 5410–5427. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, A.; Österroos, A.; Hassan, S.; Gullbo, J.; Rickardson, L.; Jarvius, M.; Nygren, P.; Fryknäs, M.; Höglund, M.; Larsson, R. Drug screen in patient cells suggests quinacrine to be repositioned for treatment of acute myeloid leukemia. Blood Cancer J. 2015, 5, e307. [Google Scholar] [CrossRef] [Green Version]
- Kuenzi, B.M.; Remsing Rix, L.L.; Kinose, F.; Kroeger, J.L.; Lancet, J.E.; Padron, E.; Rix, U. Off-target based drug repurposing opportunities for tivantinib in acute myeloid leukemia. Sci. Rep. 2019, 9, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Efferth, T. Repurposing of artemisinin-type drugs for the treatment of acute leukemia. Semin. Cancer Biol. 2021, 68, 291–312. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Chang, H.H.; Kuo, C.C.; Shiao, H.Y.; Hsieh, H.P.; Coumar, M.S. Drug repurposing for chronic myeloid leukemia: In silico and in vitro investigation of DrugBank database for allosteric Bcr-Abl inhibitors. J. Biomol. Struct. Dyn. 2017, 35, 1833–1848. [Google Scholar] [CrossRef] [PubMed]
- Sohraby, F.; Bagheri, M.; Aliyar, M.; Aryapour, H. In silico drug repurposing of FDA-approved drugs to predict new inhibitors for drug resistant T315I mutant and wild-type BCR-ABL1: A virtual screening and molecular dynamics study. J. Mol. Graph. Model 2017, 74, 234–240. [Google Scholar] [CrossRef]
- Frismantas, V.; Dobay, M.P.; Rinaldi, A.; Tchinda, J.; Dunn, S.H.; Kunz, J.; Richter-Pechanska, P.; Marovca, B.; Pail, O.; Jenni, S.; et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood 2017, 129, e26–e37. [Google Scholar] [CrossRef] [Green Version]
- Scuoppo, C.; Wang, J.; Persaud, M.; Mittan, S.K.; Basso, K.; Pasqualucci, L.; Rabadan, R.; Inghirami, G.; Grandori, C.; Bosch, F.; et al. Repurposing dasatinib for diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 2019, 116, 16981–16986. [Google Scholar] [CrossRef] [Green Version]
- Karube, K.; Enjuanes, A.; Dlouhy, I.; Jares, P.; Martin-Garcia, D.; Nadeu, F. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018, 32, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Yu, X.; Zhang, C.; Cai, Y.; Cao, Y.; Wang, S.; Shen, J. Drug Repurposing Screen Identifies Novel Classes of Drugs with Anticancer Activity in Mantle Cell Lymphoma. Comb. Chem. High Throughput Screen 2019, 22, 483–495. [Google Scholar] [CrossRef]
- Shen, M.; Zhang, Y.; Saba, N.; Austin, C.P.; Wiestner, A.; Auld, D.S. Identification of therapeutic candidates for chronic lymphocytic leukemia from a library of approved drugs. PLoS ONE 2013, 8, e75252. [Google Scholar] [CrossRef] [PubMed]
- Cooney, J.D.; Lin, A.P.; Jiang, D.; Wang, L.; Suhasini, A.N.; Myers, J.; Qiu, Z.; Wölfler, A.; Sill, H.; Aguiar, R.C.T. Synergistic Targeting of the Regulatory and Catalytic Subunits of PI3Kδ in Mature B-cell Malignancies. Clin. Cancer Res. 2018, 24, 1103–1113. [Google Scholar] [CrossRef] [Green Version]
- Chanas-Larue, A.; Villalpando-Rodriguez, G.E.; Henson, E.S.; Johnston, J.B.; Gibson, S.B. Antihistamines are synergistic with Bruton’s tyrosine kinase inhibiter ibrutinib mediated by lysosome disruption in chronic lymphocytic leukemia (CLL) cells. Leuk. Res. 2020, 96, 106423. [Google Scholar] [CrossRef]
- Mahoney, E.; Maddocks, K.; Flynn, J.; Jones, J.; Cole, S.L.; Zhang, X.; Byrd, J.C.; Johnson, A.J. Identification of endoplasmic reticulum stress-inducing agents by antagonizing autophagy: A new potential strategy for identification of anti-cancer therapeutics in B-cell malignancies. Leuk. Lymphoma 2013, 54, 2685–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez, N.; Tripathi, R.; Giro, A.; Rosich, L.; Lopez-Guerra, M.; Lopez-Oreja, I.; Playa-Albinyana, H.; Arenas, F.; Mas, J.M.; Perez-Galan, P.; et al. Systems biology drug screening identifies statins as enhancers of current therapies in chronic lymphocytic leukemia. Sci. Rep. 2020, 10, 22153. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Lotfi Shahreza, M.; Ghadiri, N.; Mousavi, S.R.; Varshosaz, J.; Green, J.R. A review of network-based approaches to drug repositioning. Brief Bioinform. 2018, 19, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Gns, H.S.; Gr, S.; Murahari, M.; Krishnamurthy, M. An update on Drug Repurposing: Re-written saga of the drug’s fate. Biomed. Pharmacother. 2019, 110, 700–716. [Google Scholar] [CrossRef]
- Glicksberg, B.S.; Li, L.; Chen, R.; Dudley, J.; Chen, B. Leveraging Big Data to Transform Drug Discovery. Methods Mol. Biol. 2019, 1939, 91–118. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Desai, R.J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdemir, E.S.; Halakou, F.; Nussinov, R.; Gursoy, A.; Keskin, O. Methods for Discovering and Targeting Druggable Protein-Protein Interfaces and Their Application to Repurposing. Methods Mol. Biol. 2019, 1903, 1–21. [Google Scholar] [CrossRef]
- Banovic, P.; Stankov, S.; Vranjes, N.; Zurkovic, O.; Capo, I.; Lalosevic, D. Drug repurposing: Mebendazole as effective antitumor agent. Are we seeing the whole story? J. Buon. 2018, 23, 1904–1911. [Google Scholar]
- Cavalla, D. Using human experience to identify drug repurposing opportunities: Theory and practice. Br. J. Clin. Pharmacol. 2019, 85, 680–689. [Google Scholar] [CrossRef]
- Pulley, J.M.; Rhoads, J.P.; Jerome, R.N.; Challa, A.P.; Erreger, K.B.; Joly, M.M.; Lavieri, R.R.; Perry, K.E.; Zaleski, N.M.; Shirey-Rice, J.K.; et al. Using What We Already Have: Uncovering New Drug Repurposing Strategies in Existing Omics Data. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 333–352. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.C.; Chen, H.H.; Gorthi, A.; Mostavi, M.; Zheng, S.; Huang, Y.; Chen, Y. Deep learning of pharmacogenomics resources: Moving towards precision oncology. Brief Bioinform. 2020, 21, 2066–2083. [Google Scholar] [CrossRef]
- Qian, T.; Zhu, S.; Hoshida, Y. Use of big data in drug development for precision medicine: An update. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 189–200. [Google Scholar] [CrossRef]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 112. [Google Scholar] [CrossRef]
- Laganà, A.; Beno, I.; Melnekoff, D.; Leshchenko, V.; Madduri, D.; Ramdas, D.; Sanchez, L.; Niglio, S.; Perumal, D.; Kidd, B.A.; et al. Precision Medicine for Relapsed Multiple Myeloma on the Basis of an Integrative Multiomics Approach. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.X.; He, Y.C.; Zhang, J.Y.; Wang, H.F.; Zhong, C.; Wang, X.T. Using Prognosis-Related Gene Expression Signature and Connectivity Map for Personalized Drug Repositioning in Multiple Myeloma. Med. Sci. Monit. 2019, 25, 3247–3255. [Google Scholar] [CrossRef] [PubMed]
- Conte, F.; Fiscon, G.; Licursi, V.; Bizzarri, D.; D’Antò, T.; Farina, L.; Paci, P. A paradigm shift in medicine: A comprehensive review of network-based approaches. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194416. [Google Scholar] [CrossRef] [PubMed]
- Nicora, G.; Vitali, F.; Dagliati, A.; Geifman, N.; Bellazzi, R. Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools. Front. Oncol. 2020, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Tanoli, Z.; Alam, Z.; Ianevski, A.; Wennerberg, K.; Vähä-Koskela, M.; Aittokallio, T. Interactive visual analysis of drug-target interaction networks using Drug Target Profiler, with applications to precision medicine and drug repurposing. Brief Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, K.; Chen, C.Z.; Marshall, R.E.; Mihatov, N.; Choi, Y.; Nguyen, D.T.; Southall, N.; Chen, K.G.; Park, J.K.; Zheng, W. Advancing precision medicine with personalized drug screening. Drug Discov. Today 2019, 24, 272–278. [Google Scholar] [CrossRef]
- Velez, G.; Bassuk, A.G.; Colgan, D.; Tsang, S.H.; Mahajan, V.B. Therapeutic drug repositioning using personalized proteomics of liquid biopsies. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Pineiro-Yanez, E.; Reboiro-Jato, M.; Gomez-Lopez, G.; Perales-Paton, J.; Troule, K.; Rodriguez, J.M.; Tejero, H.; Shimamura, T.; Lopez-Casas, P.P.; Carretero, J.; et al. PanDrugs: A novel method to prioritize anticancer drug treatments according to individual genomic data. Genome Med. 2018, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef]

 : one study and
: one study and  ,
,  ,
,  ,
,  ,
,  , or
, or  : the proteomics data of an individual person,
: the proteomics data of an individual person,  : patients with aggressive disease,
: patients with aggressive disease,  : patients with indolent disease,
: patients with indolent disease,  : healthy donors,
: healthy donors,  : patients,
: patients,  : patients with other diseases and
: patients with other diseases and  : patients of the disease of interest). 2. Data classification into subgroups with different biological questions that could be compared in the proteome level, e.g., comparison of proteomics data between patients with different stages of the disease, between healthy and patients or between patients with the disease of interest and patients from other diseases; selection of the appropriate group. 3. Comparative analysis of the proteomics data from the selected subgroup, e.g., healthy donors vs. patients. (
: patients of the disease of interest). 2. Data classification into subgroups with different biological questions that could be compared in the proteome level, e.g., comparison of proteomics data between patients with different stages of the disease, between healthy and patients or between patients with the disease of interest and patients from other diseases; selection of the appropriate group. 3. Comparative analysis of the proteomics data from the selected subgroup, e.g., healthy donors vs. patients. (  and
and  : color gradiation of the differences (small → big)). 4. Translation of the different proteome levels into deregulated pathways, in which these proteins are involved (
: color gradiation of the differences (small → big)). 4. Translation of the different proteome levels into deregulated pathways, in which these proteins are involved (  : color gradation of the differences (negative → positive)). 5. Linking of deregulated pathways to FDA-approved drugs documented through their mechanism of action. 6. Drug selection and in vitro assessment of the drugs both in the appropriate subgroup and in a subgroup control, e.g., cell cultures from healthy donors (A) and patients (B); evaluation of the pharmaceutical effect. 7. Determination of their molecular mechanisms of action, both expected and unexpected.
: color gradation of the differences (negative → positive)). 5. Linking of deregulated pathways to FDA-approved drugs documented through their mechanism of action. 6. Drug selection and in vitro assessment of the drugs both in the appropriate subgroup and in a subgroup control, e.g., cell cultures from healthy donors (A) and patients (B); evaluation of the pharmaceutical effect. 7. Determination of their molecular mechanisms of action, both expected and unexpected.
 : one study and
: one study and  ,
,  ,
,  ,
,  ,
,  , or
, or  : the proteomics data of an individual person,
: the proteomics data of an individual person,  : patients with aggressive disease,
: patients with aggressive disease,  : patients with indolent disease,
: patients with indolent disease,  : healthy donors,
: healthy donors,  : patients,
: patients,  : patients with other diseases and
: patients with other diseases and  : patients of the disease of interest). 2. Data classification into subgroups with different biological questions that could be compared in the proteome level, e.g., comparison of proteomics data between patients with different stages of the disease, between healthy and patients or between patients with the disease of interest and patients from other diseases; selection of the appropriate group. 3. Comparative analysis of the proteomics data from the selected subgroup, e.g., healthy donors vs. patients. (
: patients of the disease of interest). 2. Data classification into subgroups with different biological questions that could be compared in the proteome level, e.g., comparison of proteomics data between patients with different stages of the disease, between healthy and patients or between patients with the disease of interest and patients from other diseases; selection of the appropriate group. 3. Comparative analysis of the proteomics data from the selected subgroup, e.g., healthy donors vs. patients. (  and
and  : color gradiation of the differences (small → big)). 4. Translation of the different proteome levels into deregulated pathways, in which these proteins are involved (
: color gradiation of the differences (small → big)). 4. Translation of the different proteome levels into deregulated pathways, in which these proteins are involved (  : color gradation of the differences (negative → positive)). 5. Linking of deregulated pathways to FDA-approved drugs documented through their mechanism of action. 6. Drug selection and in vitro assessment of the drugs both in the appropriate subgroup and in a subgroup control, e.g., cell cultures from healthy donors (A) and patients (B); evaluation of the pharmaceutical effect. 7. Determination of their molecular mechanisms of action, both expected and unexpected.
: color gradation of the differences (negative → positive)). 5. Linking of deregulated pathways to FDA-approved drugs documented through their mechanism of action. 6. Drug selection and in vitro assessment of the drugs both in the appropriate subgroup and in a subgroup control, e.g., cell cultures from healthy donors (A) and patients (B); evaluation of the pharmaceutical effect. 7. Determination of their molecular mechanisms of action, both expected and unexpected.

| Repurposed Drugs | Targeted Proteins | Status of Evidence | Publication |
|---|---|---|---|
| acitretin | RXRA | in silico + experimental | [56] |
| alitretinoin | RXRA | in silico | [56] |
| aplidine | MAPK8 | in silico | [56] |
| arsenic trioxide | PML | in silico + CT PhI/II | [56] |
| auranofin | IL-1β, TNF, IL-6, thioredoxin reductase | experimental + CT PhI/II | [122] |
| azacytidine | DNA methyltransferases, DNMT1 | in silico + experimental + CT PhI/II | [56,122] |
| belimumab | BAFF | experimental | [122] |
| belinostat | HDAC8, HDAC3 | in silico | [56] |
| benoxaprofen | ALOX6 | in silico | [56] |
| bexarotene | RXRA | in silico | [56] |
| chloroquine | autophagy-related proteins | experimental | [125] |
| cladribine | RRM2B | in silico + experimental + CT PhI/II | [56] |
| clemastine | sphingosine | experimental | [124] |
| dasatinib | LCK | in silico + experimental + CT PhI/II | [56] |
| decitabine | DNMT1 | in silico | [56] |
| diclofenac | ALOX7 | in silico | [56] |
| dimercaprol | N/A | experimental | [123] |
| elomotecan | TOP1 | in silico | [56] |
| elsamitrucin | TOP1 | in silico + CT PhII | [56] |
| estramustin | MAP2 | in silico | [56] |
| etretinate | RXRA | in silico | [56] |
| gossypol | BCL2 | in silico + experimental + CT PhI/II | [56] |
| hydroxyurea | RRM2B | in silico | [56] |
| MK 1775 | WEE1 | in silico | [56] |
| nelfinavir | HIV protease | experimental | [125] |
| nintedanib | LCK | in silico | [56] |
| N-methyl-4-lle-cyclosporin | PPIA | in silico | [56] |
| oblimersen | BCL2 | in silico + PhI/II | [56] |
| paclitaxel | BCL2 | in silico | [56] |
| pazopanib | LCK | in silico + experimental | [56] |
| pentosan polysulfate | FGF2 | in silico | [56] |
| plicamycin | NA | experimental | [122] |
| podofilox | DNA topoisomerase II | experimental | [122] |
| pyroxamide | HDAC9, HDAC3 | in silico | [56] |
| rasagiline | BCL2 | in silico | [56] |
| roflumilast | PDE4 | experimental | [123] |
| simvastatin | HMGCR, LFA-1 | in silico + experimental + CT PhI | [126] |
| sucralfate | FGF4 | in silico | [56] |
| suradista | FGF3 | in silico | [56] |
| T 0128 | TOP1 | in silico | [56] |
| TA 270 | ALOX5 | in silico | [56] |
| talmapimod | MAPK13 | in silico | [56] |
| tin mesoporphyrin | HMOX1/2 | in silico | [56] |
| tretinoin | RXRA | in silico + CT PhI | [56] |
| triapine | RRM2B | in silico | [56] |
| tributyrin | HDAC7 | in silico | [56] |
| tributyrin | HDAC3 | in silico | [56] |
| valproic acid | ALDH5A1 | in silico + experimental + CT PhI/II | [56] |
| vemurafenib | FGR | in silico | [56] |
| Stage | Drugs | Targets |
|---|---|---|
| Proposed for CLL patients | BORTEZOMIB | AIFM1, BCL2, HNRNPH1, TRAF2 |
| PACLITAXEL | BCL2, STMN1, TRAF2 | |
| DOCETAXEL | BCL2, TRAF2 | |
| ERIBULIN MESYLATE | BCL2, TRAF2 | |
| VALPROIC ACID | BCL2, FAS, HDAC8 | |
| VENETOCLAX | BCL2, TRAF2 | |
| VITAMIN E | ALOX5, BCL2 | |
| Proposed for advanced CLL patients | ABEMACICLIB | CDK4, CDK6, CDKN2A, JUN |
| ARSENIC TRIOXIDE | CD44, CDK4, CDKN2A, JUN, JUP, LEF1, MAPK8, RAP1A | |
| BOSUTINIB | CAV1, CDK4, CDKN2A, ITGB1, RAP1A | |
| MIDOSTAURIN | NPM1, RAP1A, ZAP70 | |
| PACLITAXEL | CDKN2A, MAPK8, PDCD4 | |
| PALBOCICLIB | CDK4, CDK6, CDKN2A, JUN | |
| RIBOCICLIB | CDK4, CDK6, CDKN2A, JUN | |
| SIROLIMUS | CCNA1, FKBP4, MAPK8, NPM1, ZAP70 | |
| TRAMETINIB | CDKN2A, RAP1A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mavridou, D.; Psatha, K.; Aivaliotis, M. Proteomics and Drug Repurposing in CLL towards Precision Medicine. Cancers 2021, 13, 3391. https://doi.org/10.3390/cancers13143391
Mavridou D, Psatha K, Aivaliotis M. Proteomics and Drug Repurposing in CLL towards Precision Medicine. Cancers. 2021; 13(14):3391. https://doi.org/10.3390/cancers13143391
Chicago/Turabian StyleMavridou, Dimitra, Konstantina Psatha, and Michalis Aivaliotis. 2021. "Proteomics and Drug Repurposing in CLL towards Precision Medicine" Cancers 13, no. 14: 3391. https://doi.org/10.3390/cancers13143391