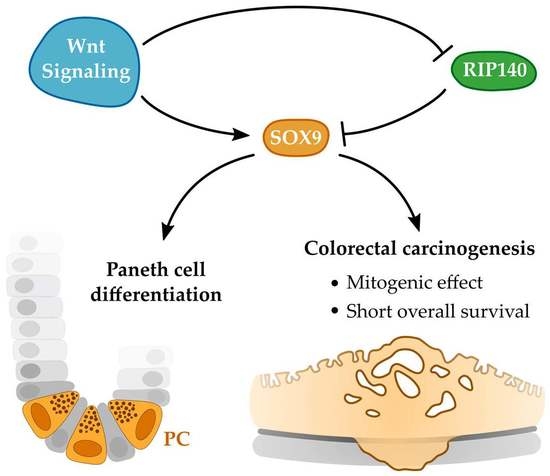
RIP140 Represses Intestinal Paneth Cell Differentiation and Interplays with SOX9 Signaling in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Organoids
2.3. Real-Time Quantitative PCR (RT-qPCR)
2.4. Histological and Immunostaining Analysis
2.5. Cell Culture and Transfections
2.6. GST Pull-Down Assay
2.7. DuoLink Proximity Ligation Assay
2.8. DNA Microarray and RNA Sequencing Analysis
2.9. Statistical Analysis
3. Results
3.1. Rip140 Inhibits Sox9 Expression and Paneth Cell Lineage in Mouse Intestine
3.2. Repression of Sox9 Expression and Paneth Cell Differentiation by Rip140 in Organoid Culture
3.3. RIP140 Is Transcriptionally Repressed by the Wnt Pathway in Human Colorectal Cell Lines
3.4. RIP140 Interacts with SOX9 and Represses Its Activity
3.5. RIP140 and SOX9 Are Opposite Prognosis Markers in CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 1974, 141, 461–479. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Johansson, M.E.; Gustafsson, J.K.; Bergström, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal. Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Gribble, F.M.; Reimann, F. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 2019, 15, 226–237. [Google Scholar] [CrossRef]
- Schneider, C.; O’Leary, C.E.; Locksley, R.M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 2019, 19, 584–593. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011, 9, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, Ö.H.; Katajisto, P.; Lamming, D.W.; Gültekin, Y.; Bauer-Rowe, K.E.; Sengupta, S.; Birsoy, K.; Dursun, A.; Yilmaz, V.O.; Selig, M.; et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 2012, 486, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Adolph, T.E.; Tomczak, M.F.; Niederreiter, L.; Ko, H.-J.; Böck, J.; Martinez-Naves, E.; Glickman, J.N.; Tschurtschenthaler, M.; Hartwig, J.; Hosomi, S.; et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013, 503, 272–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [Green Version]
- Noah, T.K.; Donahue, B.; Shroyer, N.F. Intestinal development and differentiation. Exp. Cell Res. 2011, 317, 2702–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastide, P.; Darido, C.; Pannequin, J.; Kist, R.; Robine, S.; Marty-Double, C.; Bibeau, F.; Scherer, G.; Joubert, D.; Hollande, F.; et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007, 178, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Van Es, J.H.; Jay, P.; Gregorieff, A.; van Gijn, M.E.; Jonkheer, S.; Hatzis, P.; Thiele, A.; van den Born, M.; Begthel, H.; Brabletz, T.; et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005, 7, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Van Es, J.H.; Van Gijn, M.E.; Riccio, O.; van den Born, M.; Vooijs, M.; Begthel, H.; Cozijnsen, M.; Robine, S.; Winton, D.J.; Radtke, F.; et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 2005, 435, 959–963. [Google Scholar] [CrossRef]
- Pellegrinet, L.; Rodilla, V.; Liu, Z.; Chen, S.; Koch, U.; Espinosa, L.; Kaestner, K.H.; Kopan, R.; Lewis, J.; Radtke, F. Dll1- and Dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 2011, 140, 1230–1240.e7. [Google Scholar] [CrossRef] [Green Version]
- Wong, V.W.Y.; Stange, D.E.; Page, M.E.; Buczacki, S.; Wabik, A.; Itami, S.; van de Wetering, M.; Poulsom, R.; Wright, N.A.; Trotter, M.W.B.; et al. Lrig1 controls intestinal stem cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012, 14, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Haramis, A.P.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012, 143, 1518–1529.e7. [Google Scholar] [CrossRef]
- Blache, P.; Van de Wetering, M.; Duluc, I.; Domon, C.; Berta, P.; Freund, J.-N.; Clevers, H.; Jay, P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004, 166, 37–47. [Google Scholar] [CrossRef]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [Green Version]
- Fodde, R. The APC gene in colorectal cancer. Eur. J. Cancer 2002, 38, 867–871. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Matheu, A.; Collado, M.; Wise, C.; Manterola, L.; Cekaite, L.; Tye, A.J.; Canamero, M.; Bujanda, L.; Schedl, A.; Cheah, K.S.E.; et al. Oncogenicity of the developmental transcription factor Sox. Cancer Res. 2012, 72, 1301–1315. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Garcia, E.; Lopez, L.; Aldaz, P.; Arevalo, S.; Aldaregia, J.; Egaña, L.; Bujanda, L.; Cheung, M.; Sampron, N.; Garcia, I.; et al. SOX9-regulated cell plasticity in colorectal metastasis is attenuated by rapamycin. Sci. Rep. 2016, 6, 32350. [Google Scholar] [CrossRef] [Green Version]
- Cavaillès, V.; Dauvois, S.; L’Horset, F.; Lopez, G.; Hoare, S.; Kushner, P.J.; Parker, M.G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995, 14, 3741–3751. [Google Scholar] [CrossRef]
- Ghosh, S.; Thakur, M.K. Tissue-specific expression of receptor-interacting protein in aging mouse. Age 2008, 30, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nautiyal, J. Transcriptional coregulator RIP140: An essential regulator of physiology. J. Mol. Endocrinol. 2017, 58, R147–R158. [Google Scholar] [CrossRef] [Green Version]
- Augereau, P.; Badia, E.; Carascossa, S.; Castet, A.; Fritsch, S.; Harmand, P.-O.; Jalaguier, S.; Cavaillès, V. The nuclear receptor transcriptional coregulator RIP140. Nucl. Recept. Signal. 2006, 4, nrs-04024. [Google Scholar] [CrossRef]
- Christian, M.; Tullet, J.M.A.; Parker, M.G. Characterization of four autonomous repression domains in the corepressor receptor interacting protein. J. Biol. Chem. 2004, 279, 15645–15651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapierre, M.; Bonnet, S.; Bascoul-Mollevi, C.; Ait-Arsa, I.; Jalaguier, S.; Del Rio, M.; Plateroti, M.; Roepman, P.; Ychou, M.; Pannequin, J.; et al. RIP140 increases APC expression and controls intestinal homeostasis and tumorigenesis. J. Clin. Invest. 2014, 124, 1899–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, X.; Osland, J.; Gerber, S.J.; Luan, C.; Delfino, K.; Goodwin, L.; Yuan, R. Deletion of Nrip1 extends female mice longevity, increases autophagy, and delays cell senescence. J. Gerontol. Ser. A 2018, 73, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Marjou, F.E.; Janssen, K.-P.; Chang, B.H.-J.; Li, M.; Hindie, V.; Chan, L.; Louvard, D.; Chambon, P.; Metzger, D.; Robine, S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004, 39, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Schewe, M.; Sacchetti, A.; Feijtel, D.; van de Geer, W.S.; Teeuwssen, M.; Sleddens, H.F.; Joosten, R.; van Royen, M.E.; van de Werken, H.J.G.; et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-kit signaling. Cell Rep. 2018, 24, 2312–2328.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kress, E.; Rezza, A.; Nadjar, J.; Samarut, J.; Plateroti, M. The frizzled-related sFRP2 gene is a target of thyroid hormone receptor alpha1 and activates beta-catenin signaling in mouse intestine. J. Biol. Chem. 2009, 284, 1234–1241. [Google Scholar] [CrossRef] [Green Version]
- Marisa, L.; Reyniès, A.D.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.-C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLOS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortazar, A.R.; Torrano, V.; Martín-Martín, N.; Caro-Maldonado, A.; Camacho, L.; Hermanova, I.; Guruceaga, E.; Lorenzo-Martín, L.F.; Caloto, R.; Gomis, R.R.; et al. CANCERTOOL: A visualization and representation interface to exploit cancer datasets. Cancer Res 2018, 78, 6320–6328. [Google Scholar] [CrossRef] [Green Version]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Tong, K.; Zhao, Y.; Balasubramanian, I.; Yap, G.S.; Ferraris, R.P.; Bonder, E.M.; Verzi, M.P.; Gao, N. Paneth cell multipotency induced by notch activation following injury. Cell Stem Cell 2018, 23, 46–59.e5. [Google Scholar] [CrossRef] [Green Version]
- Hoverter, N.P.; Waterman, M.L. A Wnt-fall for gene regulation: Repression. Sci. Signal. 2008, 1, pe43. [Google Scholar] [CrossRef]
- Blauwkamp, T.A.; Chang, M.V.; Cadigan, K.M. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J. 2008, 27, 1436–1446. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Goetsch, P.D.; Pike, J.W. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene expression. Mol. Endocrinol. 2012, 26, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Harley, V.R.; Clarkson, M.J.; Argentaro, A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) Box 9. Endocr. Rev. 2003, 24, 466–487. [Google Scholar] [CrossRef]
- Zhou, G.; Zheng, Q.; Engin, F.; Munivez, E.; Chen, Y.; Sebald, E.; Krakow, D.; Lee, B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19004–19009. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Chiang, C.-I.; Labhart, P.; Zhao, Y.; Yang, J.; Mistretta, T.-A.; Henning, S.J.; Maity, S.N.; Mori-Akiyama, Y. Context-specific role of SOX9 in NF-Y mediated gene regulation in colorectal cancer cells. Nucleic Acids Res. 2015, 43, 6257–6269. [Google Scholar] [CrossRef] [PubMed]
- Leung, V.Y.L.; Gao, B.; Leung, K.K.H.; Melhado, I.G.; Wynn, S.L.; Au, T.Y.K.; Dung, N.W.F.; Lau, J.Y.B.; Mak, A.C.Y.; Chan, D.; et al. SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 2011, 7, e1002356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, E.; Shen, J. The roles and functions of Paneth cells in Crohn’s disease: A critical review. Cell Prolif. 2021, 54, e12958. [Google Scholar] [CrossRef]
- Symonds, D.A. Paneth cell metaplasia in diseases of the colon and rectum. Arch. Pathol. 1974, 97, 343–347. [Google Scholar]
- Singh, R.; Balasubramanian, I.; Zhang, L.; Gao, N. Metaplastic Paneth cells in extra-intestinal mucosal niche indicate a link to microbiome and inflammation. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Gu, M.; Li, M. Plasticity of Paneth cells and their ability to regulate intestinal stem cells. Stem Cell Res. Ther. 2020, 11, 349. [Google Scholar] [CrossRef] [PubMed]
- Prévostel, C.; Blache, P. The dose-dependent effect of SOX9 and its incidence in colorectal cancer. Eur. J. Cancer Oxf. Engl. 2017, 86, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Medina, M.; Avendaño-Félix, M.; Lizárraga-Verdugo, E.; Bermúdez, M.; Romero-Quintana, J.G.; Ramos-Payan, R.; Ruíz-García, E.; López-Camarillo, C. SOX9 stem-cell factor: Clinical and functional relevance in cancer. J. Oncol. 2019, 2019, 6754040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahmanto, A.S.; Swartling, F.J.; Sangfelt, O. Targeting SOX9 for degradation to inhibit chemoresistance, metastatic spread, and recurrence. Mol. Cell Oncol. 2017, 4, e1252871. [Google Scholar] [CrossRef] [Green Version]
- Jana, S.; Krishna, B.M.; Singhal, J.; Horne, D.; Awasthi, S.; Salgia, R.; Singhal, S.S. SOX9: The master regulator of cell fate in breast cancer. Biochem. Pharmacol. 2020, 174, 113789. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gleizes, A.; Triki, M.; Bonnet, S.; Baccari, N.; Jimenez-Dominguez, G.; Covinhes, A.; Pirot, N.; Blache, P.; Yuan, R.; Győrffy, B.; et al. RIP140 Represses Intestinal Paneth Cell Differentiation and Interplays with SOX9 Signaling in Colorectal Cancer. Cancers 2021, 13, 3192. https://doi.org/10.3390/cancers13133192
Gleizes A, Triki M, Bonnet S, Baccari N, Jimenez-Dominguez G, Covinhes A, Pirot N, Blache P, Yuan R, Győrffy B, et al. RIP140 Represses Intestinal Paneth Cell Differentiation and Interplays with SOX9 Signaling in Colorectal Cancer. Cancers. 2021; 13(13):3192. https://doi.org/10.3390/cancers13133192
Chicago/Turabian StyleGleizes, Antoine, Mouna Triki, Sandrine Bonnet, Naomi Baccari, Gabriel Jimenez-Dominguez, Aurélie Covinhes, Nelly Pirot, Philippe Blache, Rong Yuan, Balázs Győrffy, and et al. 2021. "RIP140 Represses Intestinal Paneth Cell Differentiation and Interplays with SOX9 Signaling in Colorectal Cancer" Cancers 13, no. 13: 3192. https://doi.org/10.3390/cancers13133192