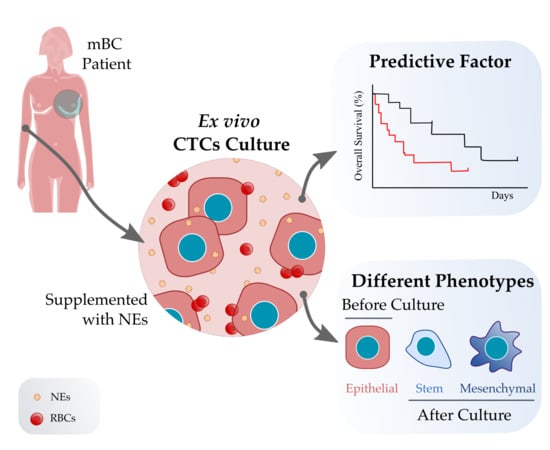
Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. CTC Isolation and Ex Vivo Culture
2.3. CTC Enumeration
2.4. Immunofluorescence Staining, Fluorescence Microscopy and Confocal Microscopy Analysis
2.5. RNA Extraction and Gene Expression Analysis
2.6. Statistical Analysis
3. Results
3.1. Patient’s and Samples Characteristics
3.2. CTC Short-Term Culture from Metastatic Breast Cancer Patients
3.2.1. CTC Culture Characterization
3.2.2. Gene Expression Analysis of Paired Samples before and after Culture
3.3. CTC Cultivability for More than 23 Days Predicts the Patient’s Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef]
- Redig, A.J.; McAllister, S.S. Breast cancer as a systemic disease: A view of metastasis. J. Intern. Med. 2013, 48, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyfried, T.N.; Huysentruyt, L.C. On the Origin of Cancer Metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Yan, W.-T.; Cui, X.; Chen, Q.; Li, Y.-F.; Cui, Y.-H.; Wang, Y.; Jiang, J. Circulating tumor cell status monitors the treatment responses in breast cancer patients: A meta-analysis. Sci. Rep. 2017, 7, srep43464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alix-Panabières, C.; Bartkowiak, K.; Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 2016, 10, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Maheswaran, S.; Haber, D.A. Ex Vivo Culture of CTCs: An Emerging Resource to Guide Cancer Therapy. Cancer Res. 2015, 75, 2411–2415. [Google Scholar] [CrossRef] [Green Version]
- Kowalik, A.; Kowalewska, M.; Góźdź, S. Current approaches for avoiding the limitations of circulating tumor cells detection methods—implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 2017, 185, 58–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nat. Cell Biol. 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Van Der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016, 7, 62754–62766. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The Identification and Characterization of Breast Cancer CTCs Competent for Brain Metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and Characterization of a Cell Line from Human Circulating Colon Cancer Cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef] [Green Version]
- Fina, E.; Reduzzi, C.; Motta, R.; Di Cosimo, S.; Bianchi, G.; Martinetti, A.; Wechsler, J.; Cappelletti, V.; Daidone, M.G. Did Circulating Tumor Cells Tell us all they Could? The Missed Circulating Tumor Cell Message in Breast Cancer. Int. J. Biol. Markers 2015, 30, 429–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; McGill, J.R.; Stanton, K.; Kassner, J.D.; Choudhury, S.; Schlegel, R.; Sauna, Z.E.; Pohlmann, P.R.; Agarwal, S. Efficient Propagation of Circulating Tumor Cells: A First Step for Probing Tumor Metastasis. Cancers 2020, 12, 2784. [Google Scholar] [CrossRef]
- Bobek, V.; Matkowski, R.; Gürlich, R.; Grabowski, K.; Szelachowska, J.; Lischke, R.; Schützner, J.; Harustiak, T.; Pazdro, A.; Rzechonek, A.; et al. Cultivation of circulating tumor cells in esophageal cancer. Folia Histochem. Cytobiol. 2014, 52, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Bobek, V.; Kacprzak, G.; Rzechonek, A.; Kolostova, K. Detection and cultivation of circulating tumor cells in malignant pleural mesothelioma. Anticancer Res. 2014, 34, 2565–2570. [Google Scholar]
- Kolostova, K.; Matkowski, R.; Gürlich, R.; Grabowski, K.; Soter, K.; Lischke, R.; Schutzner, J.; Bobek, V. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology 2015, 68, 1095–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cegan, M.; Kolostova, K.; Matkowski, R.; Broul, M.; Schraml, J.; Fiutowski, M.; Bobek, V. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 7164–7171. [Google Scholar] [PubMed]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Shiratsuchi, H.; Lin, J.; Chen, G.; Reddy, R.M.; Azizi, E.; Fouladdel, S.; Chang, A.C.; Lin, L.; Jiang, H.; et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014, 5, 12383–12397. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shiratsuchi, H.; Palanisamy, N.; Nagrath, S.; Ramnath, N. Expanded Circulating Tumor Cells from a Patient with ALK- Positive Lung Cancer Present with EML4-ALK Rearrangement Along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J. Thorac. Oncol. 2017, 12, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Que, Z.; Luo, B.; Zhou, Z.; Dong, C.; Jiang, Y.; Wang, L.; Shi, Q.; Tian, J. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019, 19, 21. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, G.; Hochmair, M.; Rath, B.; Klameth, L.; Zeillinger, R. Small cell lung cancer: Circulating tumor cells of extended stage patients express a mesenchymal-epithelial transition phenotype. Cell Adhes. Migr. 2016, 10, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Kolostova, K.; Broul, M.; Schraml, J.; Cegan, M.; Matkowski, R.; Fiutowski, M.; Bobek, V. Circulating tumor cells in localized prostate cancer: Isolation, cultivation in vitro and relationship to T-stage and Gleason score. Anticancer Res. 2014, 34, 3641–3646. [Google Scholar]
- Khoo, B.L.; Lee, S.C.; Kumar, P.; Tan, T.Z.; Warkiani, M.E.; Ow, S.G.; Nandi, S.; Lim, C.T.; Thiery, J.P. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 2015, 6, 15578–15593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef] [Green Version]
- Koch, C.; Kuske, A.; Joosse, S.A.; Yigit, G.; Sflomos, G.; Thaler, S.; Smit, D.J.; Werner, S.; Borgmann, K.; Gärtner, S.; et al. Characterization of circulating breast cancer cells with tumorigenic and metastatic capacity. EMBO Mol. Med. 2020, 12, e11908. [Google Scholar] [CrossRef]
- Sheng, W.; Ogunwobi, O.; Chen, T.; Zhang, J.; George, T.J.; Liu, C.; Fan, Z.H. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 2014, 14, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brungs, D.; Minaei, E.; Piper, A.-K.; Perry, J.; Splitt, A.; Carolan, M.; Ryan, S.; Wu, X.J.; Corde, S.; Tehei, M.; et al. Establishment of novel long-term cultures from EpCAM positive and negative circulating tumour cells from patients with metastatic gastroesophageal cancer. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Soler, A.; Cayrefourcq, L.; Mazard, T.; Babayan, A.; Lamy, P.-J.; Assou, S.; Assenat, E.; Pantel, K.; Alix-Panabières, C. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci. Rep. 2018, 8, 15931. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Cochonneau, D.; Cadé, M.; Jubelin, C.; Heymann, M.-F.; Heymann, D. Circulating Tumor Cell-Derived Pre-Clinical Models for Personalized Medicine. Cancers 2018, 11, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [Green Version]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Tosoni, D.; Di Fiore, P.P.; Pece, S. Functional Purification of Human and Mouse Mammary Stem Cells. Methods Mol. Biol. 2012, 916, 59–79. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhang, J.; Sun, B.; Zhengfeng, Y.; Jinling, Z.; Liu, S.; Sui, G.; Yin, Z. Microfluidic chip for isolation of viable circulating tumor cells of hepatocellular carcinoma for their culture and drug sensitivity assay. Cancer Biol. Ther. 2016, 17, 1177–1187. [Google Scholar] [CrossRef] [Green Version]
- Klameth, L.; Rath, B.; Hochmaier, M.; Moser, D.; Redl, M.; Mungenast, F.; Gelles, K.; Ulsperger, E.; Zeillinger, R.; Hamilton, G. Small cell lung cancer: Model of circulating tumor cell tumorospheres in chemoresistance. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vishnoi, M.; Peddibhotla, S.; Yin, W.; Scamardo, A.T.; George, G.C.; Hong, D.S.; Marchetti, D. The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci. Rep. 2015, 5, 17533. [Google Scholar] [CrossRef] [Green Version]
- Soeda, A.; Park, M.; Lee, D.; Mintz, A.; Androutsellis-Theotokis, A.; McKay, R.D.; Engh, J.; Iwama, T.; Kunisada, T.; Kassam, A.B.; et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1α. Oncogene 2009, 28, 3949–3959. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, T.; Tardito, S. Cell Culture Medium Formulation and Its Implications in Cancer Metabolism. Trends Cancer 2019, 5, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ule, N.; Abuín-Redondo, C.; Costa, C.; Piñeiro, R.; Pereira-Veiga, T.; Martínez-Pena, I.; Hurtado, P.; López-López, R.; de la Fuente, M.; Dávila-Ibáñez, A. Corrigendum to “Nanoemulsions to support ex vivo cell culture of breast cancer circulating tumor cells” [Materials Today Chemistry 16 (2020) 100265]. Mater. Today Chem. 2020, 18, 100370. [Google Scholar] [CrossRef]
- Avagliano, A.; Ruocco, M.R.; Aliotta, F.; Belviso, I.; Accurso, A.; Masone, S.; Montagnani, S.; Arcucci, A. Mitochondrial Flexibility of Breast Cancers: A Growth Advantage and a Therapeutic Opportunity. Cells 2019, 8, 401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nath, A.; Chan, C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci. Rep. 2016, 6, srep18669. [Google Scholar] [CrossRef] [Green Version]
- Antalis, C.; Uchida, A.; Buhman, K.; Siddiqui, R.A. Migration of MDA-MB-231 breast cancer cells depends on the availability of exogenous lipids and cholesterol esterification. Clin. Exp. Metastasis 2011, 28, 733–741. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Che, J.; Yu, V.; Dhar, M.; Renier, C.; Matsumoto, M.; Heirich, K.; Garon, E.B.; Goldman, J.; Rao, J.; Sledge, G.W.; et al. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget 2016, 7, 12748–12760. [Google Scholar] [CrossRef] [Green Version]
- Oeyen, S.; Liégeois, V.; De Laere, B.; Buys, A.; Strijbos, M.; Dirix, P.; Meijnders, P.; Vermeulen, P.; Van Laere, S.; Dirix, L. Automated enumeration and phenotypic characterization of CTCs and tdEVs in patients with metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Toss, A.; Mu, Z.; Fernandez, S.; Cristofanilli, M. CTC enumeration and characterization: Moving toward personalized medicine. Ann. Transl. Med. 2014, 2, 108. [Google Scholar] [PubMed]
- D’Oronzo, S.; Lovero, D.; Palmirotta, R.; Stucci, L.S.; Tucci, M.; Felici, C.; Cascardi, E.; Giardina, C.; Cafforio, P.; Silvestris, F. Dissection of major cancer gene variants in subsets of circulating tumor cells in advanced breast cancer. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kulasinghe, A.; Kapeleris, J.; Cooper, C.; Warkiani, M.E.; O’Byrne, K.; Punyadeera, C. Phenotypic Characterization of Circulating Lung Cancer Cells for Clinically Actionable Targets. Cancers 2019, 11, 380. [Google Scholar] [CrossRef] [Green Version]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Pawlikowska, P.; Farace, F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhao, X.; Cheng, C.; Li, N.; Liu, Y.; Cao, Y. The implications of signaling lipids in cancer metastasis. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monaco, M.E. Fatty acid metabolism in breast cancer subtypes. Oncotarget 2017, 8, 29487–29500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty acid synthase promotes breast cancer metastasis by mediating changes in fatty acid metabolism. Oncol. Lett. 2020, 21, 1. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Perry, C.; Warkiani, M.E.; Blick, T.; Davies, A.; O’Byrne, K.; Thompson, E.W.; Nelson, C.C.; Vela, I.; Punyadeera, C. Short term ex-vivo expansion of circulating head and neck tumour cells. Oncotarget 2016, 7, 60101–60109. [Google Scholar] [CrossRef] [Green Version]
- Kapeleris, J.; Kulasinghe, A.; Warkiani, M.E.; Oleary, C.; Vela, I.; Leo, P.; Sternes, P.; O’Byrne, K.; Punyadeera, C. Ex vivo culture of circulating tumour cells derived from non-small cell lung cancer. Transl. Lung Cancer Res. 2020, 9, 1795–1809. [Google Scholar] [CrossRef]
- Wang, R.; Chu, G.C.; Mrdenovic, S.; Annamalai, A.A.; Hendifar, A.E.; Nissen, N.N.; Tomlinson, J.S.; Lewis, M.; Palanisamy, N.; Tseng, H.-R.; et al. Cultured circulating tumor cells and their derived xenografts for personalized oncology. Asian J. Urol. 2016, 3, 240–253. [Google Scholar] [CrossRef] [Green Version]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Xin, Y.; Li, K.; Yang, M.; Tan, Y. Fluid Shear Stress Induces EMT of Circulating Tumor Cells via JNK Signaling in Favor of Their Survival during Hematogenous Dissemination. Int. J. Mol. Sci. 2020, 21, 8115. [Google Scholar] [CrossRef]
- Peake, B.F.; Eze, S.M.; Yang, L.; Castellino, R.C.; Nahta, R. Erratum: Growth differentiation factor 15 mediates epithelial mesenchymal transition and invasion of breast cancers through IGF-1R-FoxM1 signaling. Oncotarget 2020, 11, 4074–4075. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.; Dwivedi, S.; Roy, D.; Khokhar, M.; Purohit, P.; Vishnoi, J.; Pareek, P.; Sharma, S.; Sharma, P.; Misra, S. Growth differentiation factor 15 and its role in carcinogenesis: An update. Growth Factors 2019, 37, 190–207. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S.; Kawabata, T.; Aoyagi, K.; Yokozaki, H.; Sasaki, H. Gene expression and pathway analysis of CTNNB1 in cancer and stem cells. World J. Stem Cells 2016, 8, 384–395. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chang, S.-C.; Lam, S.; Ramos, R.I.; Tran, K.; Ohe, S.; Salomon, M.P.; Bhagat, A.A.S.; Lim, C.T.; Fischer, T.D.; et al. Prospective Molecular Profiling of Circulating Tumor Cells from Patients with Melanoma Receiving Combinatorial Immunotherapy. Clin. Chem. 2019, 66, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Hissa, B.; Győrffy, B.; Jann, J.-C.; Yang, C.; Reissfelder, C.; Schölch, S. Characterization of Stem-like Circulating Tumor Cells in Pancreatic Cancer. Diagnostics 2020, 10, 305. [Google Scholar] [CrossRef]
- Li, L.; Li, W. Epithelial–mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol. Ther. 2015, 150, 33–46. [Google Scholar] [CrossRef]
- Morel, A.-P.; Lièvre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of Breast Cancer Stem Cells through Epithelial-Mesenchymal Transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Wilson, M.M.; Weinberg, R.A.; Lees, J.A.; Guen, V.J. Emerging Mechanisms by which EMT Programs Control Stemness. Trends Cancer 2020, 6, 775–780. [Google Scholar] [CrossRef]
- Pereira-Veiga, T.; Martínez-Fernández, M.; Abuin, C.; Piñeiro, R.; Cebey, V.; Cueva, J.; Palacios, P.; Blanco, C.; Muinelo-Romay, L.; Abalo, A.; et al. CTCs Expression Profiling for Advanced Breast Cancer Monitoring. Cancers 2019, 11, 1941. [Google Scholar] [CrossRef] [Green Version]
- Reduzzi, C.; Vismara, M.; Gerratana, L.; Silvestri, M.; De Braud, F.; Raspagliesi, F.; Verzoni, E.; Di Cosimo, S.; Locati, L.D.; Cristofanilli, M.; et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin. Cancer Biol. 2020, 60, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Chinen, L.; Mello, C.; Abdallah, E.; Ocea, L.; Buim, M.; Mingues, N.; Junior, J.G.; Fanelli, M.; Paterlini, P. Isolation, detection, and immunomorphological characterization of circulating tumor cells (CTCs) from patients with different types of sarcoma using isolation by size of tumor cells: A window on sarcoma-cell invasion. OncoTargets Ther. 2014, 7, 1609–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emerson, T.; Kirby, M.; Bethel, K.; Kolatkar, A.; Luttgen, M.; O’Hara, S.; Newton, P.; Kuhn, P. Fourier-ring descriptor to characterize rare circulating cells from images generated using immunofluorescence microscopy. Comput. Med. Imaging Graph. 2015, 40, 70–87. [Google Scholar] [CrossRef]
- Lazar, D.C.; Cho, E.H.; Luttgen, M.S.; Metzner, T.J.; Uson, M.L.; Torrey, M.; Gross, M.; Kuhn, P. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line. Phys. Biol. 2012, 9, 016002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boshuizen, R.; Kuhn, P.; Heuvel, M.V.D. Circulating tumor cells in non-small cell lung carcinoma. J. Thorac. Dis. 2012, 4, 456–458. [Google Scholar]
- Adams, D.L.; Martin, S.S.; Alpaugh, R.K.; Charpentier, M.; Tsai, S.; Bergan, R.C.; Ogden, I.M.; Catalona, W.; Chumsri, S.; Tang, C.-M.; et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc. Natl. Acad. Sci. USA 2014, 111, 3514–3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef]
- Kang, Q.; Henry, N.L.; Paoletti, C.; Jiang, H.; Vats, P.; Chinnaiyan, A.M.; Hayes, D.F.; Merajver, S.D.; Rae, J.M.; Tewari, M. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 2016, 49, 1354–1360. [Google Scholar] [CrossRef]
- Aguirre, L.A.; Montalbán-Hernández, K.; Avendaño-Ortiz, J.; Marín, E.; Lozano, R.; Toledano, V.; Sánchez-Maroto, L.; Terrón, V.; Valentín, J.; Pulido, E.; et al. Tumor stem cells fuse with monocytes to form highly invasive tumor-hybrid cells. OncoImmunology 2020, 9, 1773204. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.P.; Singh, T.; Kumar, P.; Sharma, P.; Kaur, H.; Sharma, S.; Singh, S.; Kumar, S.; Mehta, K. Metabolic Adaptations in Cancer Stem Cells. Front. Oncol. 2020, 10, 1010. [Google Scholar] [CrossRef]
- Zhao, J.; Zhi, Z.; Wang, C.; Xing, H.; Song, G.; Yu, X.; Zhu, Y.; Wang, X.; Zhang, X.; Di, Y. Exogenous lipids promote the growth of breast cancer cells via CD36. Oncol. Rep. 2017, 38, 2105–2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.-O.; Berenguer-Llergo, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nat. Cell Biol. 2017, 541, 41–45. [Google Scholar] [CrossRef]
- Feng, W.; Wilkins, O.; Bang, S.; Ung, M.; Li, J.; An, J.; Del Genio, C.; Canfield, K.; DiRenzo, J.; Wells, W.; et al. CD36-Mediated Metabolic Rewiring of Breast Cancer Cells Promotes Resistance to HER2-Targeted Therapies. Cell Rep. 2019, 29, 3405–3420.e5. [Google Scholar] [CrossRef] [Green Version]
- Dicato, M.; Plawny, L.; Diederich, M. Anemia in cancer. Ann. Oncol. 2010, 21, vii167–vii172. [Google Scholar] [CrossRef]
- Lang, E.; Bissinger, R.; Qadri, S.M.; Lang, F. Suicidal death of erythrocytes in cancer and its chemotherapy: A potential target in the treatment of tumor-associated anemia. Int. J. Cancer 2017, 141, 1522–1528. [Google Scholar] [CrossRef] [Green Version]
- Lominadze, D.; Dean, W.L. Involvement of fibrinogen specific binding in erythrocyte aggregation. FEBS Lett. 2002, 517, 41–44. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Veiga, T.; Abreu, M.; Robledo, D.; Matias-Guiu, X.; Santacana, M.; Sánchez, L.; Cueva, J.; Palacios, P.; Abdulkader, I.; López-López, R.; et al. CTCs-derived xenograft development in a triple negative breast cancer case. Int. J. Cancer 2018, 144, 2254–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Han, H.; Liu, L.; Duan, Y.; Yang, X.; Ma, C.; Zhu, Y.; Han, J.; Li, X.; Chen, Y. CD36 plays a critical role in proliferation, migration and tamoxifen-inhibited growth of ER-positive breast cancer cells. Oncogenesis 2018, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lallo, A.; Schenk, M.W.; Frese, K.K.; Blackhall, F.; Dive, C. Circulating tumor cells and CDX models as a tool for preclinical drug development. Transl. Lung Cancer Res. 2017, 6, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Category | n | % |
|---|---|---|
| Age | ||
| ≤57 | 18 | 51.43 |
| >57 | 17 | 48.57 |
| Tumor stage | ||
| IV | 35 | 100 |
| ER status | ||
| Positive | 21 | 60 |
| Negative | 12 | 34.29 |
| PR status | ||
| Positive | 15 | 42.86 |
| Negative | 18 | 51.43 |
| HER2 status | ||
| Positive | 8 | 22.86 |
| Negative | 26 | 74.29 |
| Metastasis location | ||
| Bone | 26 | 74.29 |
| Visceral | 32 | 91.43 |
| Bone & Viceral | 23 | 65.71 |
| Number of metastatic sites | ||
| 1 | 4 | 11.43 |
| 2 | 13 | 37.14 |
| ≥3 | 18 | 51.43 |
| Therapy | ||
| Chemotherapy | 26 | 74.29 |
| CDKi + ET | 6 | 17.14 |
| ET | 2 | 5.71 |
| Lines of therapy | ||
| Basal | 14 | 40 |
| 1 Line | 7 | 20 |
| ≥2 Lines | 14 | 40 |
| Progression | 31 | 88.57 |
| Exitus | 24 | 68.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmona-Ule, N.; González-Conde, M.; Abuín, C.; Cueva, J.F.; Palacios, P.; López-López, R.; Costa, C.; Dávila-Ibáñez, A.B. Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications. Cancers 2021, 13, 2668. https://doi.org/10.3390/cancers13112668
Carmona-Ule N, González-Conde M, Abuín C, Cueva JF, Palacios P, López-López R, Costa C, Dávila-Ibáñez AB. Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications. Cancers. 2021; 13(11):2668. https://doi.org/10.3390/cancers13112668
Chicago/Turabian StyleCarmona-Ule, Nuria, Miriam González-Conde, Carmen Abuín, Juan F. Cueva, Patricia Palacios, Rafael López-López, Clotilde Costa, and Ana Belén Dávila-Ibáñez. 2021. "Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications" Cancers 13, no. 11: 2668. https://doi.org/10.3390/cancers13112668