Microfluidic Impedimetric Cell Regeneration Assay to Monitor the Enhanced Cytotoxic Effect of Nanomaterial Perfusion
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Culture
2.2. Off-Chip Cytotoxicity Assays Using Standard Cell Culture Conditions
2.3. Lab-on-a-Chip Fabrication
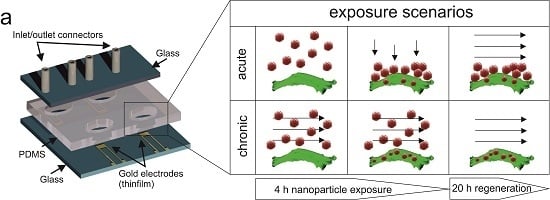
2.4. Device Preparation and on-Chip Impedimetric Regeneration Assay
3. Results and Discussion
3.1. Characterization of the on-Chip Impedance Biosensors


3.2. Toxicological Characterization of AmSil30 Nanoparticles
3.3. Comparison of Acute and Chronic NP Administration Scenarios on H441 Tumor Regeneration

3.4. Impact of Increasing Flow Rates on Tumor Regeneration during NP Administration


4. Conclusions
Supplementary Files
Supplementary File 1Author Contributions
Conflicts of Interest
References
- Moghimi, S.M.; Hunter, A.C.; Murray, J.C. Nanomedicine: Current status and future prospects. FASEB 2005, 19, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Freitas, R.A. Nanomedicine, Volume I: Basic Capabilities; Landes Bioscience: Georgetown, TX, USA, 1999. [Google Scholar]
- Poole, C.P.; Owens, F.J. Introduction to Nanotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Santamaria, A. Historical overview of nanotechnology and nanotoxicology. In Nanotoxicity: Methods and Protocols, 1st ed.; Reineke, J., Ed.; Humana Press: New York, NY, USA, 2012; Volume 926, pp. 1–12. [Google Scholar]
- Xu, Z.P.; Zeng, Q.H.; Lu, G.Q.; Yu, A.B. Inorganic nanoparticles as carriers for efficient cellular delivery. Chem. Eng. Sci. 2006, 61, 1027–1040. [Google Scholar] [CrossRef]
- Maeda, M.; Kuroda, C.S.; Shimura, T.; Tada, M.; Abe, M.; Yamamuro, S.; Sumiyama, K.; Handa, H. Magnetic carriers of iron nanoparticles coated with a functional polymer for high throughput bioscreening. J. Appl. Phys. 2006, 99. [Google Scholar] [CrossRef]
- Hilger, I.; Hergt, R.; Kaiser, W.A. Use of magnetic nanoparticle heating in the treatment of breast cancer. IEEE Proc. Nanobiotechnol. 2005, 152, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Manke, A.; Wang, L.; Rojanasakul, Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Lunov, O.; Syrovets, T.; Rocker, C.; Tron, K.; Nienhaus, G.U.; Rasche, V.; Mailander, V.; Landfester, K.; Simmet, T. Lysosomal degradation of the carboxydextran shell of coated superparamagnetic iron oxide nanoparticles and the fate of professional phagocytes. Biomaterials 2010, 31, 9015–9022. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.J.; Hutchison, G.; Christensen, F.M.; Peters, S.; Hankin, S.; Stone, V. A review of the in vivo and in vitro toxicity of silver and gold particulates: Particle attributes and biological mechanisms responsible for the observed toxicity. Crit. Rev. Toxicol. 2010, 40, 328–346. [Google Scholar] [CrossRef] [PubMed]
- Ju-Nam, Y.; Lead, J.R. Manufactured nanoparticles: An overview of their chemistry, interactions and potential environmental implications. Sci. Total Environ. 2008, 400, 396–414. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xia, T.; Nel, A.E. The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic. Biol. Med. 2008, 44, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Stone, V.; Johnston, H.; Clift, M.J. Air pollution, ultrafine and nanoparticle toxicology: Cellular and molecular interactions. IEEE Trans. Nanobiosci. 2007, 6, 331–340. [Google Scholar] [CrossRef]
- Nystrom, A.M.; Fadeel, B. Safety assessment of nanomaterials: Implications for nanomedicine. J. Controll. Release 2012, 161, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Baun, A.; Hansen, S.F. Environmental challenges for nanomedicine. Nanomedicine 2008, 3, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Bawa, R. Regulating nanomedicine—Can the fda handle it? Curr. Drug Deliv. 2011, 8, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Ahmad, M.Z.; Kazmi, I.; Akhter, S.; Afzal, M.; Gupta, G.; Sinha, V.R. Emergence of nanomedicine as cancer targeted magic bullets: Recent development and need to address the toxicity apprehension. Curr. Drug Discov. Technol. 2012, 9, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.; Walzl, A.; Unger, C.; Rosner, M.; Krupitza, G.; Hengstschlager, M.; Dolznig, H. In vitro cell migration and invasion assays. Mutat. Res. 2013, 752, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Richter, L.; Charwat, V.; Jungreuthmayer, C.; Bellutti, F.; Brueckl, H.; Ertl, P. Monitoring cellular stress responses to nanoparticles using a lab-on-a-chip. Lab Chip 2011, 11, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Yoon, T.H. Effects of ag nanoparticle flow rates on the progress of the cell cycle under continuously flowing “dynamic” exposure conditions. Bull. Korean Chem. Soc. 2014, 35, 123–128. [Google Scholar] [CrossRef]
- Mahto, S.K.; Yoon, T.H.; Rhee, S.W. A new perspective on in vitro assessment method for evaluating quantum dot toxicity by using microfluidics technology. Biomicrofluidics 2010, 4. [Google Scholar] [CrossRef] [PubMed]
- Sticker, D.; Rothbauer, M.; Charwat, V.; Steinkühler, J.; Bethge, O.; Bertagnolli, E.; Wanzenboeck, H.D.; Ertl, P. Zirconium dioxide nanolayer passivated impedimetric sensors for cell-based assays. Sens. Actuat. B: Chem. 2015, 213, 35–44. [Google Scholar] [CrossRef]
- Docter, D.; Bantz, C.; Westmeier, D.; Galla, H.J.; Wang, Q.; Kirkpatrick, J.C.; Nielsen, P.; Maskos, M.; Stauber, R.H. The protein corona protects against size- and dose-dependent toxicity of amorphous silica nanoparticles. Beilstein J. Nanotechnol. 2014, 5, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Docter, D.; Strieth, S.; Westmeier, D.; Hayden, O.; Gao, M.; Knauer, S.K.; Stauber, R.H. No king without a crown—Impact of the nanomaterial-protein corona on nanobiomedicine. Nanomedicine 2015, 10, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned—Challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Mergel, K.; Weng, A.; Frech, S.; Gilabert-Oriol, R.; Bachran, D.; Melzig, M.F.; Fuchs, H. Real time monitoring of the cell viability during treatment with tumor-targeted toxins and saponins using impedance measurement. Biosens. Bioelectron. 2012, 35, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.J.; Yu, J.J.; Xiao, L.; Tang, J.C.O.; Zhang, Y.; Wang, P.; Yang, M. Impedance studies of bio-behavior and chemosensitivity of cancer cells by micro-electrode arrays. Biosens. Bioelectron. 2009, 24, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liu, Z.; Yang, M.; Mak, A. Nanoporous membrane-based cell chip for the study of anti-cancer drug effect of retinoic acid with impedance spectroscopy. Talanta 2009, 80, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.F.; Wang, W.; Duan, W.; Zheng, F.; Sinclair, A.J.; Chatwin, C.R. Bioimpedance analysis for the characterization of breast cancer cells in suspension. IEEE Trans. Biomed. Eng. 2012, 59, 2321–2329. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kim, H.S.; Frazier, A.B.; Chen, Z.G.; Shin, D.M.; Han, A. Whole-cell impedance analysis for highly and poorly metastatic cancer cells. J. Microelectromech. Syst. 2009, 18, 808–817. [Google Scholar]
- Srinivasaraghavan, V.; Strobl, J.; Agah, M. Bioimpedance rise in response to histone deacetylase inhibitor is a marker of mammary cancer cells within a mixed culture of normal breast cells. Lab Chip 2012, 12, 5168–5179. [Google Scholar] [CrossRef] [PubMed]
- Samuel, S.P.; Jain, N.; O’Dowd, F.; Paul, T.; Kashanin, D.; Gerard, V.A.; Gun’ko, Y.K.; Prina-Mello, A.; Volkov, Y. Multifactorial determinants that govern nanoparticle uptake by human endothelial cells under flow. Nanomedicine 2012, 7, 2943–2956. [Google Scholar]
- Freese, C.; Schreiner, D.; Anspach, L.; Bantz, C.; Maskos, M.; Unger, R.E.; Kirkpatrick, C.J. In vitro investigation of silica nanoparticle uptake into human endothelial cells under physiological cyclic stretch. Part. Fibre Toxicol. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Charwat, V.; Joksch, M.; Sticker, D.; Purtscher, M.; Rothbauer, M.; Ertl, P. Monitoring cellular stress responses using integrated high-frequency impedance spectroscopy and time-resolved elisa. Analyst 2014, 139, 5271–5282. [Google Scholar] [CrossRef] [PubMed]
- Charwat, V.; Rothbauer, M.; Tedde, S.F.; Hayden, O.; Bosch, J.J.; Muellner, P.; Hainberger, R.; Ertl, P. Monitoring dynamic interactions of tumor cells with tissue and immune cells in a lab-on-a-chip. Anal. Chem. 2013, 85, 11471–11478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, F.; Nordin, A.N.; Tarbell, J.; Voiculescu, I. Toxicity studies using mammalian cells and impedance spectroscopy method. Sens. Bio-Sens. Res. 2015, 3, 112–121. [Google Scholar] [CrossRef]
- Park, S.; Choi, J.; Kim, S. Measurement of cell-substrate impedance and characterization of cancer cell growth kinetics with mathematical model. Int. J. Precis. Eng. Manuf. 2015, 16, 1859–1866. [Google Scholar] [CrossRef]
- Koppenhofer, D.; Susloparova, A.; Docter, D.; Stauber, R.H.; Ingebrandt, S. Monitoring nanoparticle induced cell death in h441 cells using field-effect transistors. Biosens. Bioelectron. 2013, 40, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.T.; Jeon, N.L.; Huang, S.; Kane, R.S.; Wargo, C.J.; Choi, I.S.; Ingber, D.E.; Whitesides, G.M. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proc. Natl. Acad. Sci. USA 2000, 97, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Ostuni, E.; Qian, X.P.; McDonald, J.C.; Jiang, X.Y.; LeDuc, P.; Wu, M.H.; Ingber, D.E.; Whitesides, G.M. Topographical micropatterning of poly(dimethylsiloxane) using laminar flows of liquids in capillaries. Adv. Mater. 2001, 13, 570–574. [Google Scholar] [CrossRef]
- Bransky, A.; Korin, N.; Levenberg, S. Experimental and theoretical study of selective protein deposition using focused micro laminar flows. Biomed. Microdevices 2008, 10, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lei, L.; Ni, X.F.; Shi, J.; Chen, Y. Patterning bio-molecules for cell attachment at single cell levels in pdms microfluidic chips. Microelectron. Eng. 2009, 86, 1462–1464. [Google Scholar] [CrossRef]
- Fang, Y. Label-free biosensors for cell biology. Int. J. Electrochem. 2011, 2011. [Google Scholar] [CrossRef]
- Mahto, S.K.; Charwat, V.; Ertl, P.; Rothen-Rutishauser, B.; Rhee, S.W.; Sznitman, J. Microfluidic platforms for advanced risk assessments of nanomaterials. Nanotoxicology 2015, 9, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, M.; Wartmann, D.; Charwat, V.; Ertl, P. Recent advances and future applications of microfluidic live-cell microarrays. Biotechnol. Adv. 2015, 33, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Wartmann, D.; Rothbauer, M.; Kuten, O.; Barresi, C.; Visus, C.; Felzmann, T.; Ertl, P. Automated, miniaturized and integrated quality control-on-chip (qc-on-a-chip) for advanced cell therapy applications. Front. Mater. 2015, 2. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rothbauer, M.; Praisler, I.; Docter, D.; Stauber, R.H.; Ertl, P. Microfluidic Impedimetric Cell Regeneration Assay to Monitor the Enhanced Cytotoxic Effect of Nanomaterial Perfusion. Biosensors 2015, 5, 736-749. https://doi.org/10.3390/bios5040736
Rothbauer M, Praisler I, Docter D, Stauber RH, Ertl P. Microfluidic Impedimetric Cell Regeneration Assay to Monitor the Enhanced Cytotoxic Effect of Nanomaterial Perfusion. Biosensors. 2015; 5(4):736-749. https://doi.org/10.3390/bios5040736
Chicago/Turabian StyleRothbauer, Mario, Irene Praisler, Dominic Docter, Roland H. Stauber, and Peter Ertl. 2015. "Microfluidic Impedimetric Cell Regeneration Assay to Monitor the Enhanced Cytotoxic Effect of Nanomaterial Perfusion" Biosensors 5, no. 4: 736-749. https://doi.org/10.3390/bios5040736