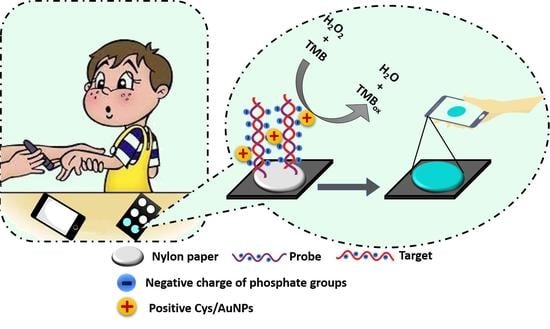
Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Apparatus
2.3. Preparation of AuNPs
2.4. Preparation of Cys/AuNPs
2.5. Paper-Based Genosensor
2.6. miRNA-21 Hybridization Process
2.7. Smartphone-Based Colorimetric Detection
3. Results and Discussion
3.1. Study of Cys/AuNPs Composites as Nanozyme
3.2. Characterization of the Nylon Membrane Disk before and after Immobilization of DNAProbe and the Study of Electrostatic Interaction of DNAprobe
3.3. Optimization of Assay
3.4. Principle of the Developed Paper-Based Genosensor for the miRNA-21 Detection
3.5. Performance of the Paper-Based Genosensor
3.6. Selectivity and Stability Study of the Paper-Based Genosensor for the miRNA-21 Detection
3.7. Application of the Paper-Based Genosensor in Real Sample
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yates, L.A.; Norbury, C.J.; Gilbert, R.J.C. The Long and Short of MicroRNA. Cell 2013, 153, 516–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Amine, A. Spectrophotometric and Electrochemical Determination of MicroRNA-155 Using Sandwich Hybridization Magnetic Beads. Anal. Lett. 2018, 51, 411–423. [Google Scholar] [CrossRef]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (Review). Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Y.; Deng, H.; Shen, W.; Gao, Z. A Highly Sensitive and Selective Electrochemical Biosensor for Direct Detection of MicroRNAs in Serum. Anal. Chem. 2013, 85, 4784–4789. [Google Scholar] [CrossRef]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef]
- El Aamri, M.; Mohammadi, H.; Amine, A. Novel Label-free Colorimetric and Electrochemical Detection for MiRNA-21 Based on the Complexation of Molybdate with Phosphate. Microchem. J. 2022, 182, 107851. [Google Scholar] [CrossRef]
- El Aamri, M.; Mohammadi, H.; Amine, A. Development of a Novel Electrochemical Sensor Based on Functionalized Carbon Black for the Detection of Guanine Released from DNA Hydrolysis. Electroanalysis 2022, 34, 573–771. [Google Scholar] [CrossRef]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef]
- Mohammadi, H.; Yammouri, G.; Amine, A. Current advances in electrochemical genosensors for detecting microRNA cancer markers. Curr. Opin. Electrochem. 2019, 16, 96–105. [Google Scholar] [CrossRef]
- Moustakim, H.; Mohammadi, H.; Amine, A. Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer. Sensors 2022, 22, 9420. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Chen, S.; Fu, P.; Lin, Y.; Ye, S.; Long, Y.; Gao, G.; Zheng, J. A persistent luminescence resonance energy transfer-based molecular beacon probe for the highly sensitive detection of microRNA in biological samples. Biosens. Bioelectron. 2022, 198, 113849. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheng, W.; Yan, Y.; Zhang, Y.; Yin, Y.; Ju, H.; Ding, S. A colorimetric biosensor for detection of attomolar microRNA with a functional nucleic acid-based amplification machine. Talanta 2016, 146, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, X.; Yang, J.; Jiang, Y.; He, N. Peroxidase-like activity of mesoporous silica encapsulated Pt nanoparticle and its application in colorimetric immunoassay. Anal. Chim. Acta 2015, 862, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.S.; Hosseini, M.; Ganjali, M.R.; Hosseinkhani, S. A novel dual-mode and label-free aptasensor based methodology for breast cancer tissue marker targeting. Sens. Actuators B Chem. 2020, 315, 128084. [Google Scholar] [CrossRef]
- Dehghani, Z.; Hosseini, M.; Mohammadnejad, J.; Ganjali, M.R. Novel colorimetric sensor based on peroxidase-like activity of chitosan-stabilized Au/Pt nanoclusters for trace lead. Anal. Methods 2019, 11, 684–690. [Google Scholar] [CrossRef]
- Dehghani, Z.; Hosseini, M.; Mohammadnejad, J.; Bakhshi, B.; Rezayan, A.H. Colorimetric aptasensor for Campylobacter jejuni cells by exploiting the peroxidase like activity of Au@Pd nanoparticles. Microchim. Acta 2018, 185, 448. [Google Scholar] [CrossRef]
- Rahimi-Nasrabadi, M.; Mizani, F.; Hosseini, M.; Keihan, A.H.; Ganjali, M.R. Detection of hydrogen peroxide and glucose by using Tb2 (MoO4)3 nanoplates as peroxidase mimics. Spectrochim. Acta 2017, 186, 82–88. [Google Scholar] [CrossRef]
- Hosseini, M.; Aghazadehb, M.; Ganjali, M.R. A facile one-pot synthesis of cobaltdoped magnetite/graphene nanocomposite as peroxidase mimetics in dopamine detection. New J. Chem. 2017, 41, 12678–12684. [Google Scholar] [CrossRef]
- Pothipor, C.; Jakmunee, J.; Bamrungsap, S.; Ounnunkad, K. An electrochemical biosensor for simultaneous detection of breast cancer clinically related microRNAs based on a gold nanoparticles/graphene quantum dots/graphene oxide film. Analyst 2021, 146, 4000–4009. [Google Scholar] [CrossRef]
- Yammouri, G.; Mandli, J.; Mohammadi, H.; Amine, A. Development of an electrochemical label-free biosensor for microRNA-125a detection using pencil graphite electrode modified with different carbon nanomaterials. J. Electroanal. Chem. 2017, 806, 75–81. [Google Scholar] [CrossRef]
- Yang, L.; Wang, J.; Lü, H.; Ni, H. Electrochemical sensor based on Prussian blue/multi-walled carbon nanotubes functionalized polypyrrole nanowire arrays for hydrogen peroxide and microRNA detection. Microchim. Acta 2021, 188, 25. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Roy, S.; Singh, P.; Ziyauddin, K.; Amit, J. 2D MoS2-based nanomaterials for therapeutic, bioimaging, and biosensing applications. Small 2019, 15, 1803706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, S.; Zhang, K.; Zhu, L.; Dianping, T. ZIF-8-assisted NaYF4: Yb, Tm@ ZnO converter with exonuclease III-powered DNA walker for near-infrared light responsive biosensor. Anal. Chem. 2019, 92, 1470–1476. [Google Scholar] [CrossRef]
- Kong, L.; Lv, S.; Qiao, Z.; Yongcun, Y.; Jian, Z.; Sai, B. Metal-organic framework nanoreactor-based electrochemical biosensor coupled with three-dimensional DNA walker for label-free detection of microRNA. Biosens. Bioelectron. 2022, 207, 114188. [Google Scholar] [CrossRef]
- Xue, Q.; Niu, X.; Liu, P.; Mengzhu, W.; Yinxian, P.; Hongbing, P.; Xin, L. Analyte-triggered citrate-stabilized Au nanoparticle aggregation with accelerated peroxidase-mimicking activity for catalysis-based colorimetric sensing of arsenite. Sens. Actuators B Chem. 2021, 334, 129650. [Google Scholar] [CrossRef]
- Abarghoei, S.; Fakhri, N.; Borghei, Y.S.; Hosseini, M.; Ganjali, M.R. A colorimetric paper sensor for citrate as biomarker for early stage detection of prostate cancer based on peroxidase-like activity of cysteine-capped gold nanoclusters. Spectrochim. Acta Part A 2019, 210, 251–259. [Google Scholar] [CrossRef]
- Fakhri, N.; Morteza, H.; Omid, T. Aptamer-based colorimetric determination of Pb2+ using a paper-based microfluidic platform. Anal. Meth. 2018, 10, 4438–4444. [Google Scholar] [CrossRef]
- Carrilho, E.; Martinez, A.W.; Whitesides, G.M. Understanding wax printing: A simple micropatterning process for paper-based microfluidics. Anal. Chem. 2009, 81, 7091–7095. [Google Scholar] [CrossRef]
- Songjaroen, T.; Dungchai, W.; Chailapakul, O.; Laiwattanapaisal, W. Novel, simple and low-cost alternative method for fabrication of paper-based microfluidics by wax dipping. Talanta 2011, 85, 2587–2593. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Zhang, Y.; Lin, L.; Zhou, C.; Li, S.; Zhang, L.; Li, J. Low-cost fabrication of paperbased microfluidic devices by one step plotting. Anal. Chem. 2012, 84, 6331–6335. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Kotera, K.; Suzuki, K.; Citterio, D. Inkjet-printed paper fluidic immunochemical sensing device. Anal. Bioanal. Chem. 2010, 398, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Sones, C.L.; Katis, I.N.; He, P.J.W.; Mills, B.; Namiq, M.F.; Shardlow, P.; Ibsen, M.; Eason, R.W. Laser-induced photo-polymerisation for creation of paper-based fluidic devices. Lab Chip 2014, 14, 4567–4574. [Google Scholar] [CrossRef] [PubMed]
- OuYang, L.; Wang, C.; Du, F.; Zheng, T.; Liang, H. Electrochromatographic separations of multicomponent metal complexes on a microfluidic paper-based device with a simplified photolithography. RSC Adv. 2014, 4, 1093–1101. [Google Scholar] [CrossRef]
- Krishnan, T.; Wang, H.-N.; Vo-Dinh, T. Smartphone-Based Device for Colorimetric Detection of MicroRNA Biomarkers Using Nanoparticle-Based Assay. Sensors 2021, 21, 8044. [Google Scholar] [CrossRef]
- Georgia, I.S.; Nicole, H.; Philip, H.E.G. Factors influencing the surface functionalization of citrate stabilized gold nanoparticles with cysteamine, 3-mercaptopropionic acid or l-Selenocystine for sensor applications. Chemosensors 2020, 8, 80. [Google Scholar]
- Lou-Franco, J.; Das, B.; Elliott, C.; Cuong, C. Gold nanozymes: From concept to biomedical applications. Nano-Micro Lett. 2021, 13, 10. [Google Scholar] [CrossRef]
- Yuhana Ariffin, E.; Heng, L.Y.; Tan, L.L.; Abd Karim, N.H.; Hasbullah, S.A. A highly sensitive impedimetric DNA biosensor based on hollow silica microspheres for label-free determination of E coli. Sensors 2020, 20, 1279. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Xu, Z.; Duan, Y.; Zhu, Y.; Ou, M.; Xu, X. Immobilization of tyrosinase on polyacrylonitrile beads: Biodegradation of phenol from aqueous solution and the relevant cytotoxicity assessment. RSC Adv. 2017, 7, 28114–28123. [Google Scholar] [CrossRef] [Green Version]
- Fakhri, N.; Abarghoei, S.; Dadmehr, M.; Hosseini, M.; Sabahi, H.; Ganjali, M.R. Paper based colorimetric detection of miRNA-21 using Ag/Pt nanoclusters. Spectrochim. Acta A 2020, 227, 117529. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Na, H.; Lee, S.; Kim, W. Advanced graphene oxide-based paper sensor for colorimetric detection of miRNA. Microchim. Acta 2022, 189, 35. [Google Scholar] [CrossRef] [PubMed]







| Nucleic Acid | Sequence (5′-3′) |
|---|---|
| Target miRNA-21 | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| DNAprobe (complementary sequence of miRNA-21): | 5′-AAATCAACATCAGTCTGATAAGCTA-3′ |
| MiRNA-146 (non-complementary oligonucleotide) | 5′-UGAGAACUGAAUUCCAUGGGUU-3’ |
| MiRNA-125a (non-complementary oligonucleotide) | 5′-UCCCUGAGACCCUUUAACCUGUGA-3’ |
| Strategies | Target | Solid Substrate | Regression Equation | Linear Range | Limit of Detection | Ref. |
|---|---|---|---|---|---|---|
| miRNA detection based on peroxidase mimetic activity of DNA-Ag/Pt NCs | miR-21 | Whatman filter | y = 0.06x + 147.48 | 10–1000 pM | 4.1 pM | [41] |
| miRNA detection based on silver NPs aggregation | miR-21 | In solution | Y = 0.355 Logx − 0.067 | 1–300 nM | - | [36] |
| Paper-based sensor for miRNAs by combining GONET and the target-recycled signal amplification strategy | miR-122 | Whatman filter | - | - | 0.52 nM | [42] |
| miRNA detection based on the generation of molybdophosphate complex coupled with DNA hydrolysis | miR-21 | 96-well microplates | Y = 82.3 − 11.5 Logx | 1–50,000 pM | 0.6 pM | [8] |
| miRNA detection-based peroxidase-mimetic activity of cysteamine-capped gold nanoparticles | miR-21 | Nylon paper | Y = 0.080x + 13.846 | 1–1000 nM | 0.5 pM | This work |
| Sample | Original [miR-21] (pM) | Spiked [miR-21] (pM) | Founded [miR-21] (pM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| 1 | 0 | 10.0 | 9.6 | 96.0 | 4.7 |
| 2 | 0 | 50.0 | 48.4 | 96.8 | 3.9 |
| 3 | 0 | 500.0 | 488.2 | 97.6 | 6.3 |
| 4 | 0 | 1000 | 900 | 90.0 | 7.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aamri, M.E.; Mohammadi, H.; Amine, A. Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles. Biosensors 2023, 13, 74. https://doi.org/10.3390/bios13010074
Aamri ME, Mohammadi H, Amine A. Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles. Biosensors. 2023; 13(1):74. https://doi.org/10.3390/bios13010074
Chicago/Turabian StyleAamri, Maliana El, Hasna Mohammadi, and Aziz Amine. 2023. "Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles" Biosensors 13, no. 1: 74. https://doi.org/10.3390/bios13010074