The Regulatory Role of Key Metabolites in the Control of Cell Signaling
Abstract
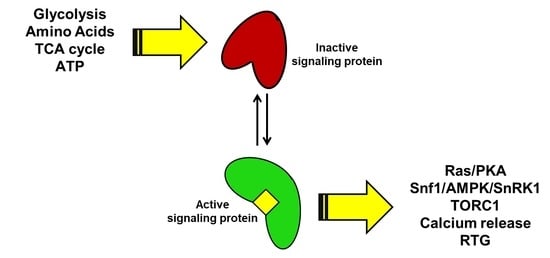
:1. Introduction
2. The Biochemistry of the Protein-Metabolite Interaction (PMI)
3. The Main Pathways of Nutrient Sensing and Cellular Signaling
3.1. Protein-Metabolite Interactions Controlling the Ras/PKA Pathway
3.2. Protein-Metabolite Interactions Controlling SNF1/AMPK/SnRK1 Pathway
3.3. Protein-Metabolite Interactions Controlling TOR Activity in Yeast and Mammalian Cells
3.4. Yeast Mitochondrial Retrograde Response Is Influenced by Protein-Metabolite Interactions
3.5. Protein-Metabolite Interactions Controlling Calcium Release
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kitano, H. Biological robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Foundations of Systems Biology; MIT Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Friedlander, T.; Mayo, A.E.; Tlusty, T.; Alon, U. Evolution of Bow-Tie Architectures in Biology. PLoS Comput. Biol. 2015, 11, e1004055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelling, J.; Sauer, U.; Szallasi, Z.; Doyle, F.J.; Doyle, J. Robustness of Cellular Functions. Cell 2004, 118, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busti, S.; Coccetti, P.; Alberghina, L.; Vanoni, M. Glucose signaling-mediated coordination of cell growth and cell cycle in saccharomyces cerevisiae. Sensors 2010, 10, 6195–6240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewald, J.C. How yeast coordinates metabolism, growth and division. Curr. Opin. Microbiol. 2018, 45, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Diether, M.; Sauer, U. Towards detecting regulatory Protein-metabolite interactions. Curr. Opin. Microbiol. 2017, 39, 16–23. [Google Scholar] [CrossRef]
- Diether, M.; Nikolaev, Y.; Allain, F.H.; Sauer, U. Systematic mapping of protein-metabolite interactions in central metabolism of Escherichia coli. Mol. Syst. Biol. 2019, 15, e9008. [Google Scholar] [CrossRef]
- Peeters, K.; Van Leemputte, F.; Fischer, B.; Bonini, B.M.; Quezada, H.; Tsytlonok, M.; Haesen, D.; Vanthienen, W.; Bernardes, N.; Gonzalez-Blas, C.B.; et al. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Zhang, C.S.; Hawley, S.A.; Zong, Y.; Li, M.; Wang, Z.; Gray, A.; Ma, T.; Cui, J.; Feng, J.W.; Zhu, M.; et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 2017, 548, 112–116. [Google Scholar] [CrossRef]
- Piazza, I.; Kochanowski, K.; Cappelletti, V.; Fuhrer, T.; Noor, E.; Sauer, U.; Picotti, P. A map of protein-metabolite interactions reveals principles of chemical communication. Cell 2018, 172, 358–372. [Google Scholar] [CrossRef] [Green Version]
- Eisenmesser, E.Z.; Millet, O.; Labeikovsky, W.; Korzhnev, D.M.; Wolf-Watz, M.; Bosco, D.A.; Skalicky, J.J.; Kay, L.E.; Kern, D. Intrinsic dynamics of an enzyme underlies catalysis. Nature 2005, 438, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lee, S.; Jeon, A.; Choi, J.M.; Lee, H.S.; Hohng, S.; Kim, H.S. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nat. Chem. Biol. 2013, 9, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianni, S.; Dogan, J.; Jemth, P. Distinguishing induced fit from conformational selection. Biophys. Chem. 2014, 189, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Koshland, D.E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA 1958, 44, 98–104. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Kumar, S.; Tsai, C.-J.; Nussinov, R. Folding funnels and binding mechanisms. Protein Eng. Des. Sel. 1999, 12, 713–720. [Google Scholar] [CrossRef] [Green Version]
- Weikl, T.R.; Paul, F. Conformational selection in protein binding and function. Protein Sci. 2014, 23, 1508–1518. [Google Scholar] [CrossRef] [Green Version]
- Kirschner, K.; Eigen, M.; Bittman, R.; Voigt, B. The binding of nicotinamide-adenine dinucleotide to yeast D-glyceraldheyde-3-phosphate dehydrogenase: Temperature-jump relaxation studies on the mechanism of an allosteric enzyme. Proc. Natl. Acad. Sci. USA 1966, 56, 1661–1667. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.A. Role of induced fit in enzyme specificity: A molecular forward/reverse switch. J. Biol. Chem. 2008, 283, 26297–26301. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.M.; Craik, C.S. Trapping moving targets with small molecules. Science 2009, 324, 213–215. [Google Scholar] [CrossRef] [Green Version]
- Wagner, J.R.; Lee, C.T.; Durrant, J.D.; Malmstrom, R.D.; Feher, V.A.; Amaro, R.E. Emerging computational methods for the rational discovery of allosteric drugs. Chem. Rev. 2016, 116, 6370–6390. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-J.; Kumar, S.; Ma, B.; Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999, 8, 1181–1190. [Google Scholar] [CrossRef] [Green Version]
- Alberghina, L.; Coccetti, P.; Orlandi, I. Systems biology of the cell cycle of Saccharomyces cerevisiae: From network mining to system-level properties. Biotechnol. Adv. 2009, 27, 960–978. [Google Scholar] [CrossRef]
- Alberghina, L.; Mavelli, G.; Drovandi, G.; Palumbo, P.; Pessina, S.; Tripodi, F.; Coccetti, P.; Vanoni, M. Cell growth and cell cycle in Saccharomyces cerevisiae: Basic regulatory design and protein-protein interaction network. Biotechnol. Adv. 2012, 30, 52–72. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef] [Green Version]
- Knorre, D.; Sokolov, S.; Zyrina, A. How do yeast sense mitochondrial dysfunction? Microb. Cell 2016, 3, 532–539. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, F.; Nicastro, R.; Reghellin, V.; Coccetti, P. Post-translational modifications on yeast carbon metabolism: Regulatory mechanisms beyond transcriptional control. Biochim. Biophys. Acta 2015, 1850, 620–627. [Google Scholar] [CrossRef]
- Steyfkens, F.; Zhang, Z.; Van Zeebroeck, G.; Thevelein, J.M. Multiple transceptors for macro- and micro-nutrients control diverse cellular properties through the PKA pathway in yeast: A paradigm for the rapidly expanding world of eukaryotic nutrient transceptors up to those in human cells. Front. Pharmacol. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Ma, P.; Cauwenberg, L.; Winderickx, J.; Crauwels, M.; Teunissen, A.; Nauwelaers, D.; de Winde, J.H.; Gorwa, M.; Colavizza, D.; et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose-and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 3326–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Batlle, M.; Hirsch, J.P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998, 17, 1996–2007. [Google Scholar] [CrossRef]
- Kataoka, T.; Powers, S.; McGill, C.; Fasano, O.; Strathern, J.; Broach, J.; Wigler, M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 1984, 37, 437–445. [Google Scholar] [CrossRef]
- Jacquet, E.; Parrini, M.C.; Bernardi, A.; Martegani, E.; Parmeggiani, A. Properties of the catalytic domain of CDC25, a saccharomyces cerevisiae GDP/GTP exchange factor: Comparison of its activity on full-length and c-terminal truncated ras2 proteins. Biochem. Biophys. Res. Commun. 1994, 199, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; de Winde, J.H.; Lemaire, K.; Boles, E.; Thevelein, J.M.; Winderickx, J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 2000, 38, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 1371–1377. [Google Scholar] [CrossRef] [Green Version]
- Rolland, F.; Wanke, V.; Cauwenberg, L.; Ma, P.; Boles, E.; Vanoni, M.; de Winde, J.H.; Thevelein, J.M.; Winderickx, J. The role of hexose transport and phosphorylation in cAMP signalling in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2001, 1, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Blázquez, M.A.; Lagunas, R.; Gancedo, C.; Gancedo, J.M. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993, 329, 51–54. [Google Scholar] [CrossRef] [Green Version]
- Heinisch, J. Construction and physiological characterization of mutants disrupted in the phosphofructokinase genes of Saccharomyces cerevisiae. Curr. Genet. 1986, 11, 227–234. [Google Scholar] [CrossRef]
- Huberts, D.H.E.W.; Niebel, B.; Heinemann, M. A flux-sensing mechanism could regulate the switch between r espiration and fermentation. FEMS Yeast Res. 2012, 12, 118–128. [Google Scholar] [CrossRef] [Green Version]
- Buck, J.; Sinclair, M.L.; Schapal, L.; Cann, M.J.; Levin, L.R. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc. Natl. Acad. Sci. USA 1999, 96, 79–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Cann, M.J.; Litvin, T.N.; Iourgenko, V.; Sinclair, M.L.; Levin, L.R.; Buck, J. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 2000, 289, 625–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cann, M.J.; Hammer, A.; Zhou, J.; Kanacher, T. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J. Biol. Chem. 2003, 278, 35033–35038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steegborn, C.; Litvin, T.N.; Levin, L.R.; Buck, J.; Wu, H. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment. Nat. Struct. Mol. Biol. 2005, 12, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Saalau-Bethell, S.M.; Berdini, V.; Cleasby, A.; Congreve, M.; Coyle, J.E.; Lock, V.; Murray, C.W.; O’Brien, M.A.; Rich, S.J.; Sambrook, T.; et al. Crystal structure of human soluble adenylate cyclase reveals a Distinct, highly flexible allosteric bicarbonate binding pocket. Chem. Med. Chem. 2014, 9, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Esposito, G.; Jaiswal, B.S.; Xie, F.; Krajnc-Franken, M.A.M.; Robben, T.J.A.A.; Strik, A.M.; Kuil, C.; Philipsen, R.L.A.; van Duin, M.; Conti, M.; et al. Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect. Proc. Natl. Acad. Sci. USA 2004, 101, 2993–2998. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, C.; Hauser, A.D.; Vucic, E.A.; Bar-Sagi, D. Plasma membrane V-ATPase controls oncogenic RAS-induced macropinocytosis. Nature 2019, 576, 477–481. [Google Scholar] [CrossRef]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef] [Green Version]
- Roelofs, J.; van Haastert, P.J.M. Deducing the Origin of Soluble Adenylyl Cyclase, a Gene Lost in Multiple Lineages. Mol. Biol. Evol. 2002, 19, 2239–2246. [Google Scholar] [CrossRef] [Green Version]
- Klengel, T.; Liang, W.J.; Chaloupka, J.; Ruoff, C.; Schröppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogensen, E.G.; Janbon, G.; Chaloupka, J.; Steegborn, C.; Man, S.F.; Moyrand, F.; Klengel, T.; Pearson, D.S.; Geeves, M.A.; Buck, J.; et al. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot. Cell 2006, 5, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungbluth, M.; Mösch, H.U.; Taxis, C. Acetate regulation of spore formation is under the control of the Ras/cyclic AMP/protein kinase a pathway and carbon dioxide in Saccharomyces cerevisiae. Eukaryot. Cell 2012, 11, 1021–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkuni, K.; Hayashi, M.; Yamashita, I. Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae. Yeast 1998, 14, 623–631. [Google Scholar] [CrossRef]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor snf1/AMPK in saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Crepin, N.; Rolland, F. SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 2019, 51, 29–36. [Google Scholar] [CrossRef]
- Carlson, M.; Osmond, B.C.; Botstein, D. Mutants of yeast defective in sucrose utilization. Genetics 1981, 98, 25–40. [Google Scholar]
- Nicastro, R.; Tripodi, F.; Guzzi, C.; Reghellin, V.; Khoomrung, S.; Capusoni, C.; Compagno, C.; Airoldi, C.; Nielsen, J.; Alberghina, L.; et al. Enhanced amino acid utilization sustains growth of cells lacking Snf1/AMPK. Biochim. Biophys. Acta 2015, 1853, 1615–1625. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, F.; Castoldi, A.; Nicastro, R.; Reghellin, V.; Lombardi, L.; Airoldi, C.; Falletta, E.; Maffioli, E.; Scarcia, P.; Palmieri, L.; et al. Methionine supplementation stimulates mitochondrial respiration. Biochim. Biophys. Acta 2018, 1865, 1901–1913. [Google Scholar] [CrossRef]
- Castermans, D.; Somers, I.; Kriel, J.; Louwet, W.; Wera, S.; Versele, M.; Janssens, V.; Thevelein, J.M. Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res. 2012, 22, 1058–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, A.; Xu, X.; Carlson, M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc. Natl. Acad. Sci. USA 2011, 108, 6349–6354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, A.; Xu, X.; Carlson, M. Ptc1 protein phosphatase 2C contributes to glucose regulation of SNF1/AMP-activated protein kinase (AMPK) in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 31052–31058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, F.V.; Heath, R.; Underwood, E.; Sanders, M.J.; Carmena, D.; McCartney, R.R.; Leiper, F.C.; Xiao, B.; Jing, C.; Walker, P.A.; et al. ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase. Cell Metab. 2011, 14, 707–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrashekarappa, D.G.; McCartney, R.R.; Schmidt, M.C. Ligand binding to the AMP-activated protein kinase active site mediates protection of the activation loop from dephosphorylation. J. Biol. Chem. 2013, 288, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, L.; Orlova, M.; Maziarz, M.; Kuchin, S. Protein Kinase A Contributes to the Negative Control of Snf1 Protein Kinase in Saccharomyces cerevisiae. Eukaryot Cell 2012, 11, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Nicastro, R.; Tripodi, F.; Gaggini, M.; Castoldi, A.; Reghellin, V.; Nonnis, S.; Tedeschi, G.; Coccetti, P. Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 24715–24726. [Google Scholar] [CrossRef] [Green Version]
- Rubenstein, E.M.; McCartney, R.R.; Zhang, C.; Shokat, K.M.; Shirra, M.K.; Arndt, K.M.; Schmidt, M.C. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 2008, 283, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Hawley, S.A.; Boudeau, J.; Reid, J.L.; Mustard, K.J.; Udd, L.; Mäkelä, T.P.; Alessi, D.R.; Hardie, D.G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003, 2, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Hawley, S.A.; Pan, D.A.; Mustard, K.J.; Ross, L.; Bain, J.; Edelman, A.M.; Frenguelli, B.G.; Hardie, D.G. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005, 2, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Carling, D.; Zammit, V.A.; Hardie, D.G. A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett. 1987, 223, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Zong, H.; Ren, J.M.; Young, L.H.; Pypaert, M.; Mu, J.; Birnbaum, M.J.; Shulman, G.I. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Natl. Acad. Sci. USA 2002, 99, 15983–15987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suter, M.; Riek, U.; Tuerk, R.; Schlattner, U.; Wallimann, T.; Neumann, D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 2006, 281, 32207–32216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, B.; Sanders, M.J.; Underwood, E.; Heath, R.; Mayer, F.V.; Carmena, D.; Jing, C.; Walker, P.A.; Eccleston, J.F.; Haire, L.F.; et al. Structure of mammalian AMPK and its regulation by ADP. Nature 2011, 472, 230–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, S.P.; Helps, N.R.; Cohen, P.T.W.; Hardie, D.G. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2Cα and native bovine protein phosphatase-2Ac. FEBS Lett. 1995, 377, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, L.; Zhou, X.E.; Ke, J.; De Waal, P.W.; Gu, X.; Tan, M.H.E.; Wang, D.; Wu, D.; Xu, H.E.; et al. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 2015, 25, 50–66. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xin, F.-J.; Wang, J.; Hu, J.; Zhang, Y.-Y.; Wan, S.; Cao, L.-S.; Lu, C.; Li, P.; Frank Yan, S.; et al. Conserved regulatory elements in AMPK. Nature 2013, 498, E8–E10. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Guo, H.; Zhang, C.S.; Lin, S.Y.; Yin, Z.; Peng, Y.; Luo, H.; Shi, Y.; Lian, G.; Zhang, C.; et al. AMP as a low-energy charge signal autonomously initiates assembly of axin-ampk-lkb1 complex for AMPK activation. Cell Metab. 2013, 18, 546–555. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.L.; Wu, Y.Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The lysosomal v-ATPase-ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014, 20, 526–540. [Google Scholar] [CrossRef] [Green Version]
- Rago, F.; Saltzberg, D.; Allen, K.N.; Tolan, D.R. Enzyme Substrate Specificity Conferred by Distinct Conformational Pathways. J. Am. Chem. Soc. 2015, 137, 13876–13886. [Google Scholar] [CrossRef]
- McBride, A.; Ghilagaber, S.; Nikolaev, A.; Hardie, D.G. The Glycogen-Binding Domain on the AMPK β Subunit Allows the Kinase to Act as a Glycogen Sensor. Cell Metab. 2009, 9, 23–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiatrowski, H.A.; van Denderen, B.J.W.; Berkey, C.D.; Kemp, B.E.; Stapleton, D.; Carlson, M. Mutations in the Gal83 Glycogen-Binding Domain Activate the Snf1/Gal83 Kinase Pathway by a Glycogen-Independent Mechanism. Mol. Cell. Biol. 2004, 24, 352–361. [Google Scholar] [CrossRef] [Green Version]
- Emanuelle, S.; Hossain, M.I.; Moller, I.E.; Pedersen, H.L.; van de Meene, A.M.L.; Doblin, M.S.; Koay, A.; Oakhill, J.S.; Scott, J.W.; Willats, W.G.T.; et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015, 82, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Muraro, D.; Byrne, H.M.; King, J.R.; Bennett, M.J. Mathematical Modelling Plant Signalling Networks. Math. Model. Nat. Phenom. 2013, 8, 5–24. [Google Scholar] [CrossRef] [Green Version]
- Emanuelle, S.; Doblin, M.S.; Stapleton, D.I.; Bacic, A.; Gooley, P.R. Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1. Trends Plant Sci. 2016, 21, 341–353. [Google Scholar] [CrossRef]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.C.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-related protein kinasel activity and regulation of metabolic pathways by trehalose-6-phosphate1[w][OA]. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Nunes, C.; Primavesi, L.F.; Patel, M.K.; Martinez-Barajas, E.; Powers, S.J.; Sagar, R.; Fevereiro, P.S.; Davis, B.G.; Paul, M.J. Inhibition of SnRK1 by metabolites: Tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 2013, 63, 89–98. [Google Scholar] [CrossRef]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing wrinkled1[open]. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef] [Green Version]
- Shimobayashi, M.; Hall, M.N. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014, 15, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Nicastro, R.; Sardu, A.; Panchaud, N.; de Virgilio, C. The Architecture of the Rag GTPase Signaling Network. Biomolecules 2017, 7, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binda, M.; Péli-Gulli, M.P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; de Virgilio, C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ukai, H.; Araki, Y.; Kira, S.; Oikawa, Y.; May, A.I.; Noda, T. Gtr/Ego-independent TORC1 activation is achieved through a glutamine-sensitive interaction with Pib2 on the vacuolar membrane. PLoS Genet. 2018, 14, e1007334. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.; Cunningham, K.W. A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress. Mol. Biol. Cell 2015, 26, 4631–4645. [Google Scholar] [CrossRef]
- Zhang, T.; Péli-Gulli, M.P.; Yang, H.; de Virgilio, C.; Ding, J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the EGO complex to activate TORC1. Structure 2012, 20, 2151–2160. [Google Scholar] [CrossRef] [Green Version]
- Kira, S.; Tabata, K.; Shirahama-Noda, K.; Nozoe, A.; Yoshimori, T.; Noda, T. Reciprocal conversion of Gtr1 and Gtr2 nucleotidebinding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy 2014, 10, 1565–1578. [Google Scholar] [CrossRef] [Green Version]
- Bonfils, G.; Jaquenoud, M.; Bontron, S.; Ostrowicz, C.; Ungermann, C.; de Virgilio, C. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol. Cell 2012, 46, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, S.; Thevelein, J.M. The cell division cycle gene CDC60 encodes cytosolic leucyl-tRNA synthetase in Saccharomyces cerevisiae. Gene 1992, 120, 43–49. [Google Scholar] [CrossRef]
- Echols, N.; Harrison, P.; Balasubramanian, S.; Luscombe, N.M.; Bertone, P.; Zhang, Z.; Gerstein, M. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 2002, 30, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Rock, F.L.; Mao, W.; Yaremchuk, A.; Tukalo, M.; Crépin, T.; Zhou, H.; Zhang, Y.K.; Hernandez, V.; Akama, T.; Baker, S.J.; et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007, 316, 1759–1761. [Google Scholar] [CrossRef] [PubMed]
- Powis, K.; de Virgilio, C. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2016, 2, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, K.; Yonezawa, K.; Weng, Q.P.; Kozlowski, M.T.; Belham, C.; Avruch, J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998, 273, 14484–14494. [Google Scholar] [CrossRef] [Green Version]
- Menon, S.; Dibble, C.C.; Talbott, G.; Hoxhaj, G.; Valvezan, A.J.; Takahashi, H.; Cantley, L.C.; Manning, B.D. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014, 156, 771–785. [Google Scholar] [CrossRef] [Green Version]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [Green Version]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Long, X.; Lin, Y.; Ortiz-Vega, S.; Yonezawa, K.; Avruch, J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005, 15, 702–713. [Google Scholar] [CrossRef] [Green Version]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef] [Green Version]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016, 351, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, R.A.; Chantranupong, L.; Knockenhauer, K.E.; Schwartz, T.U.; Sabatini, D.M. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 2016, 536, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Parsons, A.B.; Brost, R.L.; Ding, H.; Li, Z.; Zhang, C.; Sheikh, B.; Brown, G.W.; Kane, P.M.; Hughes, T.R.; Boone, C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004, 22, 62–69. [Google Scholar] [CrossRef]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Tsun, Z.Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Wade Harper, J.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P.X. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 2013, 154, 403. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Wu, Y.; Sheen, J. TOR signaling in plants: Conservation and innovation. Development 2018, 145, dev160887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Meng, Y.; Li, X.; Zhou, Y.; Ma, L.; Fu, L.; Schwarzländer, M.; Liu, H.; Xiong, Y. Metabolite-mediated TOR signaling regulates the circadian clock in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 25395–25397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nukarinen, E.; Ngele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.G.; Hoffman, G.R.; Poulogiannis, G.; Buel, G.R.; Jang, Y.J.; Lee, K.W.; Kim, B.Y.; Erikson, R.L.; Cantley, L.C.; Choo, A.Y.; et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell 2013, 49, 172–185. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Silbermann, M.; Speiser, A.; Forieri, I.; Linster, E.; Poschet, G.; Allboje Samami, A.; Wanatabe, M.; Sticht, C.; Teleman, A.A.; et al. Sulfur availability regulates plant growth via glucose-TOR signaling. Nat. Commun. 2017, 8, e1174. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.; Kim, S.J.; Xing, A.; Schenck, C.A.; Liu, L.; Jiang, N.; Wang, J.; Last, R.L.; Brandizzi, F. Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. Elife 2019, 8, e50747. [Google Scholar] [CrossRef]
- Pronk, J.T.; Steensma, H.Y.; van Dijken, J.P. Pyruvate metabolism in saccharomyces cerevisiae. Yeast 1996, 12, 1607–1633. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Seol, D.W. The role of mitochondria in apoptosis. J. Biochem. Mol. Biol. 2008, 41, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Shadel, G.S.; Horvath, T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell 2015, 163, 560–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, X.S.; Small, W.C.; Srere, P.A.; Butow, R.A. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 38–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Butow, R.A. A Transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999, 19, 6720–6728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallstrom, T.C.; Moye-Rowley, W.S. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 37347–37356. [Google Scholar] [CrossRef] [Green Version]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The retrograde response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Upadhyay, S.; Srivastava, Y. Retrograde response by reactive oxygen/nitrogen species in plants involving different cellular organelles. Biol. Chem. 2019, 400, 979–989. [Google Scholar] [CrossRef]
- Sekito, T.; Thornton, J.; Butow, R.A. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 2000, 11, 2103–2115. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Sekito, T.; SŠpiírek, M.; Thornton, J.; Butow, R.A. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell 2003, 12, 401–411. [Google Scholar] [CrossRef]
- Zhang, F.; Pracheil, T.; Thornton, J.; Liu, Z. Adenosine Triphosphate (ATP) is a candidate signaling molecule in the mitochondria-to-nucleus retrograde response pathway. Genes (Basel) 2013, 4, 86–100. [Google Scholar] [CrossRef]
- Santulli, G.; Nakashima, R.; Yuan, Q.; Marks, A.R. Intracellular calcium release channels: An update. J. Physiol. 2017, 595, 3041–3051. [Google Scholar] [CrossRef] [Green Version]
- Berridge, M.J. The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol. Rev. 2016, 96, 1261–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clapper, D.L.; Walseth, T.F.; Dargie, P.J.; Lee, H.C. Pyridine nucleotide metabolites stimulate calcium release from sea urchin egg microsomes desensitized to inositol trisphosphate. J. Biol. Chem. 1987, 262, 9561–9568. [Google Scholar] [PubMed]
- Galione, A.; Lee, H.C.; Busa, W.B. Ca2+-induced Ca2+ release in sea urchin egg homogenates: Modulation by cyclic ADP-Ribose. Science 1991, 253, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C. Potentiation of calcium- and caffeine-induced calcium release by cyclic ADP-ribose. J. Biol. Chem. 1993, 268, 293–299. [Google Scholar] [PubMed]
- Zheng, J.; Wenzhi, B.; Miao, L.; Hao, Y.; Zhang, X.; Yin, W.; Pan, J.; Yuan, Z.; Song, B.; Ji, G. Ca2+ release induced by cADP-ribose is mediated by FKBP12.6 proteins in mouse bladder smooth muscle. Cell Calcium 2010, 47, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki-Mann, M.; Demuro, A.; Parker, I. cADPR stimulates SERCA activity in Xenopus oocytes. Cell Calcium 2009, 45, 293–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki-Mann, M.; Demuro, A.; Parker, I. Modulation of endoplasmic reticulum Ca2+ store filling by cyclic ADP-ribose promotes inositol trisphosphate (IP3)-evoked Ca2+ signals. J. Biol. Chem. 2010, 285, 25053–25061. [Google Scholar] [CrossRef] [Green Version]
- Park, D.R.; Nam, T.S.; Kim, Y.W.; Lee, S.H.; Kim, U.H. CD38-cADPR-SERCA signaling axis determines skeletal muscle contractile force in response to β-adrenergic stimulation. Cell. Physiol. Biochem. 2018, 46, 2017–2030. [Google Scholar] [CrossRef]
- Lange, I.; Penner, R.; Fleig, A.; Beck, A. Synergistic regulation of endogenous TRPM2 channels by adenine dinucleotides in primary human neutrophils. Cell Calcium 2008, 44, 604–615. [Google Scholar] [CrossRef] [Green Version]
- Wolf, B.A.; Colcaq, J.R.; Cornens, P.G.; Turk, J.; Mcdaniels, M.L. Glucose 6-Phosphate Regulates Ca2+ steady state in endoplasmic reticulum of islets a possible link in glucose-induced insulin secretion. J. Biol. Chem. 1986, 261, 16284–16287. [Google Scholar]
- Chen, P.Y.; Csutora, P.; Veyna-Burke, N.A.; Marchase, R.B. Glucose-6-phosphate and Ca2+ sequestration are mutually enhanced in microsomes from liver, brain, and heart. Diabetes 1998, 47, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.T.; Kean, W.S.; Pollard, H.B.; Verma, A.; Watson, W.D. Glucose-6-phosphate reduces calcium accumulation in rat brain endoplasmic reticulum. Front. Mol. Neurosci. 2012, 5, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kockskämper, J.; Zima, A.V.; Blatter, L.A. Modulation of sarcoplasmic reticulum Ca 2+ release by glycolysis in cat atrial myocytes. J. Physiol. 2005, 564, 697–714. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, G.H.; Lamp, S.T.; Motter, C.; Bridge, J.H.B.; Garfinkel, A.; Goldhaber, J.I. Metabolic inhibition alters subcellular calcium release patterns in rat ventricular myocytes: Implications for defective excitation-contraction coupling during cardiac ischemia and failure. Circ. Res. 2005, 96, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Cyert, M.S.; Philpott, C.C. Regulation of cation balance in Saccharomyces cerevisiae. Genetics 2013, 193, 677–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittman, J.K. Vacuolar Ca2+ uptake. Cell Calcium 2011, 50, 139–146. [Google Scholar] [CrossRef]
- Bonilla, M.; Cunningham, K.W. Calcium release and influx in yeast: TRPC and VGCC rule another kingdom. Sci. STKE 2002, 2002, PE17. [Google Scholar] [CrossRef]
- Palmer, C.P.; Zhou, X.L.; Lin, J.; Loukin, S.H.; Kung, C.; Saimi, Y. A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca2+-permeable channel in the yeast vacuolar membrane. Proc. Natl. Acad. Sci. USA 2001, 98, 7801–7805. [Google Scholar] [CrossRef] [Green Version]
- Dürr, G.; Strayle, J.; Plemper, R.; Elbs, S.; Klee, S.K.; Catty, P.; Wolf, D.H.; Rudolph, H.K. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-Associated protein degradation. Mol. Biol. Cell 1998, 9, 1149–1162. [Google Scholar] [CrossRef] [Green Version]
- Tisi, R.; Belotti, F.; Wera, S.; Winderickx, J.; Thevelein, J.M.; Martegani, E. Evidence for inositol triphosphate as a second messenger for glucose-induced calcium signalling in budding yeast. Curr. Genet. 2004, 45, 83–89. [Google Scholar] [CrossRef]
- Iida, H.; Sakaguchi, S.; Yagawa, Y.; Anraku, Y.J. Cell Cycle Control by Ca2+ in Saccharomyces Cerevisiae. J. Biol. Chem. 1990, 265, 16–22. [Google Scholar]
- Busti, S.; Mapelli, V.; Tripodi, F.; Sanvito, R.; Magni, F.; Coccetti, P.; Rocchetti, M.; Nielsen, J.; Alberghina, L.; Vanoni, M. Respiratory metabolism and calorie restriction relieve persistent endoplasmic reticulum stress induced by calcium shortage in yeast. Sci. Rep. 2016, 6, 27942. [Google Scholar] [CrossRef] [Green Version]
- Tökés-Füzesi, M.; Bedwell, D.M.; Repa, I.; Sipos, K.; Sümegi, B.; Rab, A.; Miseta, A. Hexose phosphorylation and the putative calcium channel component Mid1p are required for the hexose-induced transient elevation of cytosolic calcium response in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 44, 1299–1308. [Google Scholar] [CrossRef]
- Tisi, R.; Baldassa, S.; Belotti, F.; Martegani, E. Phospholipase C is required for glucose-induced calcium influx in budding yeast. FEBS Lett. 2002, 520, 133–138. [Google Scholar] [CrossRef]
- Fu, L.; Miseta, A.; Hunton, D.; Marchase, R.B.; Bedwell, D.M. Loss of the major isoform of phosphoglucomutase results in altered calcium homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5431–5440. [Google Scholar] [CrossRef] [Green Version]
- Aiello, D.P.; Fu, L.; Miseta, A.; Sipos, K.; Bedwell, D.M. The Ca2+ homeostasis defects in a pgm2Δ strain of Saccharomyces cerevisiae are caused by excessive vacuolar Ca2+ uptake mediated by the Ca2+-ATPase Pmc1p. J. Biol. Chem. 2004, 279, 38495–38502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, D.P.; Fu, L.; Miseta, A.; Bedwell, D.M. Intracellular glucose 1-phosphate and glucose 6-phosphate levels modulate Ca2+ homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 45751–45758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dlugai, S.; Hippler, S.; Wieczorke, R.; Boles, E. Glucose-dependent and -independent signalling functions of the yeast glucose sensor Snf3. FEBS Lett. 2001, 505, 389–392. [Google Scholar] [CrossRef]
- Feng, Y.; de Franceschi, G.; Kahraman, A.; Soste, M.; Melnik, A.; Boersema, P.J.; de Laureto, P.P.; Nikolaev, Y.; Oliveira, A.P.; Picotti, P. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 2014, 32, 1036–1044. [Google Scholar] [CrossRef]
- Li, X.; Gianoulis, T.A.; Yip, K.Y.; Gerstein, M.; Snyder, M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell 2010, 143, 639–650. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milanesi, R.; Coccetti, P.; Tripodi, F. The Regulatory Role of Key Metabolites in the Control of Cell Signaling. Biomolecules 2020, 10, 862. https://doi.org/10.3390/biom10060862
Milanesi R, Coccetti P, Tripodi F. The Regulatory Role of Key Metabolites in the Control of Cell Signaling. Biomolecules. 2020; 10(6):862. https://doi.org/10.3390/biom10060862
Chicago/Turabian StyleMilanesi, Riccardo, Paola Coccetti, and Farida Tripodi. 2020. "The Regulatory Role of Key Metabolites in the Control of Cell Signaling" Biomolecules 10, no. 6: 862. https://doi.org/10.3390/biom10060862