A High-Accuracy Detection System: Based on Transfer Learning for Apical Lesions on Periapical Radiograph
Abstract
:1. Introduction
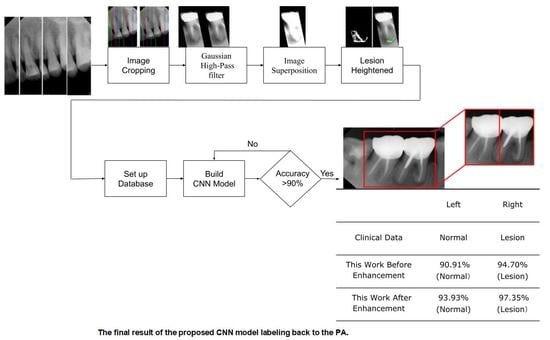
- In the image cropping preprocessing part, this study adds the adaptive threshold and angle rotation technology. Compared with the existing methods, this method significantly improves the image clarity and accuracy of a single tooth image.
- This study proposes an advanced image enhancement technique for apical lesions. It adds raw grayscale images and Gaussian high-pass filtered images to highlight the possible lesion areas and changes the color of the possible lesion area to green. Experiments show that the accuracy of the model is improved by more than 10% which proves that the proposed method is intuitive and effective.
- The innovation of this work is to realize the classification of various diseases. It can simultaneously judge a variety of different types of dental diseases (such as apical lesions, fillings, etc.), and the obtained final accuracy of the model proposed in this paper is as high as 93%. AlexNet even improves the accuracy up to 96.21% which is 4% higher than the state-of-the-art in [23].
2. Materials and Methods
2.1. Image Segmentation and Retouching
2.1.1. Vertical Cutting
2.1.2. Image Masks
2.2. Enhancing Lesion
2.2.1. Grayscale Image
2.2.2. Gaussian High Pass Filter

2.2.3. Lesion Heightened
2.3. Image Identification
2.3.1. CNN Model
2.3.2. Adjust Hyperparameter
- A.
- Initial Learning Rate
- B.
- Max Epoch
- C.
- Mini Batch Size
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Panetta, K.; Rajendran, R.; Ramesh, A.; Rao, S.P.; Agaian, S. Tufts Dental Database: A Multimodal Panoramic X-Ray Dataset for Benchmarking Diagnostic Systems. IEEE J. Biomed. Health Inform. 2022, 26, 1650–1659. [Google Scholar] [CrossRef] [PubMed]
- Karamifar, K.; Tondari, A.; Saghiri, M.A. Endodontic Periapical Lesion: An Overview on the Etiology, Diagnosis and Current Treatment Modalities. Eur. Endod. J. 2020, 5, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Luongo, R.; Faustini, F.; Vantaggiato, A.; Bianco, G.; Traini, T.; Scarano, A.; Pedullà, E.; Bugea, C. Implant Periapical Lesion: Clinical and Histological Analysis of Two Case Reports Carried Out with Two Different Approaches. Bioengineering 2022, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Sánchez, M.C.; Cabanillas-Balsera, D.; Areal-Quecuty, V.; Velasco-Ortega, E.; Martín-González, J.; Segura-Egea, J.J. Cardiovascular Diseases and Apical Periodontitis: Association Not Always Implies Causality. Med. Oral Patol. Oral Cir. Bucal 2020, 25, e652–e659. [Google Scholar] [CrossRef] [PubMed]
- Kamberi, B.; Hoxha, V.; Stavileci, M.; Dragusha, E.; Kuçi, A.; Kqiku, L. Prevalence of Apical Periodontitis and Endodontic Treatment in a Kosovar Adult Population. BMC Oral Health 2011, 11, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, P.N.R. On the Causes of Persistent Apical Periodontitis: A Review. Int. Endod. J. 2006, 39, 249–281. [Google Scholar] [CrossRef]
- Ridao-Sacie, C.; Segura-Egea, J.J.; Fernández-Palacín, A.; Bullón-Fernández, P.; Ríos-Santos, J.V. Radiological Assessment of Periapical Sta-tus Using the Periapical Index: Comparison of Periapical Radiography and Digital Panoramic Radiography. Int. Endod. J. 2007, 40, 433–440. [Google Scholar] [CrossRef]
- De Paula-Silva, F.W.G.; Wu, M.-K.; Leonardo, M.R.; da Silva, L.A.B.; Wesselink, P.R. Accuracy of Periapical Radiography and Cone-Beam Computed Tomography Scans in Diagnosing Apical Periodontitis Using Histopathological Findings as a Gold Standard. J. Endod. 2009, 35, 1009–1012. [Google Scholar] [CrossRef]
- Wallace, J.A.; Nair, M.K.; Colaco, M.F.; Kapa, S.F. A Comparative Evaluation of the Diagnostic Efficacy of Film and Digital Sensors for Detection of Simulated Periapical Lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 92, 93–97. [Google Scholar] [CrossRef]
- Chazel, J.-C.; Mafart, B. Apical Lesions. Br. Dent. J. 2004, 196, 2. [Google Scholar] [CrossRef]
- Chen, S.-L.; Chang, C.E.; Chen, C.A.; Abu, P.A.R.; Lin, T.L.; Lin, S.Y.; Chiang, W.Y.; Tu, W.C. Real-Time Image Contrast Enhancement VLSI Design for Intelligent Autonomous Vehicles. J. Imaging Sci. Technol. 2020, 64, 10504-1–10504-11. [Google Scholar] [CrossRef]
- Chi, T.-K.; Chen, H.-C.; Chen, S.-L.; Abu, P.A.R. A High-Accuracy and Power-Efficient Self-Optimizing Wireless Water Level Monitoring IoT Device for Smart City. Sensors 2021, 21, 1936. [Google Scholar] [CrossRef]
- Hsia, C.-H.; Lin, T.-Y.; Lin, J.-L.; Prasetyo, H.; Chen, S.-L.; Tseng, H.-W. System for Recommending Facial Skincare Products. Sens. Mater. 2020, 32, 3235. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Wu, C.-M.; Chen, S.-L.; Lin, T.-L.; Tseng, Y.-W. Continuous Facial Emotion Recognition Method Based on Deep Learning of Academic Emotions. Sens. Mater. 2020, 32, 3243. [Google Scholar] [CrossRef]
- Adedigba, A.P.; Adeshina, S.A.; Aibinu, A.M. Performance Evaluation of Deep Learning Models on Mammogram Classification Using Small Dataset. Bioengineering 2022, 9, 161. [Google Scholar] [CrossRef]
- Madan, P.; Singh, V.; Singh, D.P.; Diwakar, M.; Pant, B.; Kishor, A. A Hybrid Deep Learning Approach for ECG-Based Arrhythmia Classification. Bioengineering 2022, 9, 152. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, P.; Li, Y.; Mou, X.; Zhao, Z.; Chen, X.; Du, L.; Yang, T.; Zhan, Q.; Fang, Z. Prediction of Pulmonary Function Parameters Based on a Combination Algorithm. Bioengineering 2022, 9, 136. [Google Scholar] [CrossRef]
- Chen, S.-L.; Zhou, H.-S.; Chen, T.-Y.; Lee, T.-H.; Chen, C.-A.; Lin, T.-L.; Lin, N.-H.; Wang, L.-H.; Lin, S.-Y.; Chiang, W.-Y.; et al. Dental Shade Matching Method Based on Hue, Saturation, Value Color Model with Machine Learning and Fuzzy Decision. Sens. Mater. 2020, 32, 3185–3207. [Google Scholar] [CrossRef]
- Lakshmi, M.M.; Chitra, P. Tooth Decay Prediction and Classification from X-Ray Images Using Deep CNN. In Proceedings of the 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 28–30 July 2020; pp. 1349–1355. [Google Scholar]
- Lin, N.-H.; Lin, T.-L.; Wang, X.; Kao, W.-T.; Tseng, H.-W.; Chen, S.-L.; Chiou, Y.-S.; Lin, S.-Y.; Villaverde, J.; Kuo, Y.-F. Teeth Detection Algorithm and Teeth Condition Classification Based on Convolutional Neural Networks for Dental Panoramic Radiographs. J. Med. Imaging Health Inform. 2018, 8, 507–515. [Google Scholar] [CrossRef]
- Yilmaz, E.; Kayikçioğlu, T.; Kayipmaz, S. Semi-Automatic Segmentation of Apical Lesions in Cone Beam Computed Tomography Images. In Proceedings of the 2017 25th Signal Processing and Communications Applications Conference (SIU), Antalya, Turkey, 15–18 May 2017; pp. 1–4. [Google Scholar]
- Jayalakshmi, G.S.; Kumar, V.S. Performance Analysis of Convolutional Neural Network (CNN) Based Cancerous Skin Lesion Detection System. In Proceedings of the 2019 International Conference on Computational Intelligence in Data Science (ICCIDS), Chennai, India, 21–23 February 2019; pp. 1–6. [Google Scholar]
- Li, C.-W.; Lin, S.-Y.; Chou, H.-S.; Chen, T.-Y.; Chen, Y.-A.; Liu, S.-Y.; Liu, Y.-L.; Chen, C.-A.; Huang, Y.-C.; Chen, S.-L.; et al. Detection of Dental Apical Lesions Using CNNs on Periapical Radiograph. Sensors 2021, 21, 7049. [Google Scholar] [CrossRef]
- Mao, Y.-C.; Chen, T.-Y.; Chou, H.-S.; Lin, S.-Y.; Liu, S.-Y.; Chen, Y.-A.; Liu, Y.-L.; Chen, C.-A.; Huang, Y.-C.; Chen, S.-L.; et al. Caries and Restoration Detection Using Bitewing Film Based on Transfer Learning with CNNs. Sensors 2021, 21, 4613. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Tsai, H.-J. A Novel Adaptive Local Dimming Backlight Control Chip Design Based on Gaussian Distribution for Liquid Crystal Displays. J. Disp. Technol. 2016, 12, 1494–1505. [Google Scholar] [CrossRef]












| The Number of Periapical Images after Classification | |||
|---|---|---|---|
| Training Set | Validation Set | Total | |
| Normal | 332 | 83 | 415 |
| Lesion | 330 (Expanded) | 15 | 345 |
| The Number of Original Periapical Images | |||
|---|---|---|---|
| Normal | Lesion | Total | |
| Quantity | 415 | 75 | 490 |
| Hardware Platform | Version |
|---|---|
| CPU | AMD R7-5800H |
| GPU | GeForce RTX 3070 |
| DRAM | DDR4 3200 16GB |
| Software platform | Version |
| MATLAB | R2021a |
| Deep Network designer | 14.2 |
| Type | Activations | |
|---|---|---|
| 1 | Image Input | 227 × 227 × 3 |
| 2 | Convolution | 55 × 55 × 96 |
| 3 | ReLU | 55 × 55 × 96 |
| 4 | Cross Channel Normalization | 50 × 55 × 96 |
| 5 | Max pooling | 27 × 27 × 96 |
| 6 | Grouped Convolution | 27 × 27 × 256 |
| 7 | ReLU | 27 × 27 × 256 |
| 8 | Cross Channel Normalization | 27 × 27 × 256 |
| 9 | Max pooling | 13 × 13 × 256 |
| 10 | Convolution | 13 × 13 × 384 |
| 11 | ReLU | 13 × 13 × 384 |
| 12 | Grouped Convolution | 13 × 13 × 384 |
| 13 | ReLU | 13 × 13 × 384 |
| 14 | Grouped Convolution | 13 × 13 × 256 |
| 15 | ReLU | 13 × 13 × 256 |
| 16 | Max pooling | 6 × 6 × 256 |
| 17 | Fully-Connected | 1 × 1 × 4096 |
| 18 | ReLU | 1 × 1 × 4096 |
| 19 | Dropout | 1 × 1 × 4096 |
| 20 | Fully-Connected | 1 × 1 × 4096 |
| 21 | ReLU | 1 × 1 × 4096 |
| 22 | Dropout | 1 × 1 × 4096 |
| 23 | Fully-Connected | 1 × 1 × 2 |
| 24 | Softmax | 1 × 1 × 2 |
| 25 | Classification Output | 1 × 1 × 2 |
| Hyperparameters | Value |
|---|---|
| Initial Learning Rate | 0.0001 |
| Max Epoch | 50 |
| Mini Batch Size | 64 |
| Validation Frequency | 10 |
| Learning Drop Period | 3 |
| Learning Rate Drop Factor | 0.02 |
| Epoch | Iteration | Time Elapsed | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 00:00:02 | 48.44% | 53.03% | 1.4716 | 0.7940 |
| 5 | 40 | 00:00:15 | 70.31% | 81.82% | 0.5114 | 0.4379 |
| 10 | 80 | 00:00:27 | 90.62% | 85.61% | 0.2726 | 0.3277 |
| 15 | 120 | 00:00:39 | 90.62% | 88.64% | 0.2668 | 0.2648 |
| 20 | 160 | 00:00:42 | 89.06% | 90.91% | 0.2776 | 0.2422 |
| 25 | 200 | 00:01:03 | 87.50% | 91.67% | 0.3722 | 0.2230 |
| 30 | 240 | 00:01:16 | 90.62% | 93.94% | 0.1955 | 0.1787 |
| 35 | 280 | 00:01:28 | 95.31% | 95.31% | 0.1313 | 0.1883 |
| 40 | 320 | 00:01:41 | 90.62% | 95.45% | 0.2768 | 0.1585 |
| 45 | 360 | 00:01:53 | 96.88% | 95.45% | 0.0896 | 0.1424 |
| 50 | 400 | 00:02:05 | 93.72% | 96.21% | 0.1520 | 0.1201 |
| Target Class | ||||
|---|---|---|---|---|
| Category Name | Lesion | Normal | Subtotal | |
| Output Class | Lesion | 49.2% | 3.0% | 94.2% |
| Normal | 0.8% | 47.0% | 98.4% | |
| subtotal | 98.5% | 93.9% | 96.2% | |
| Tooth Position in Figure 13 | Left | Right |
|---|---|---|
| Clinical Data | Normal | Lesion |
| This Work Before Enhancement | 90.91% Normal | 94.70% Lesion |
| This Work After Enhancement | 93.93% Normal | 97.35% Lesion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuo, Y.; Lin, W.-M.; Chen, T.-Y.; Chan, M.-L.; Chang, Y.-S.; Lin, Y.-R.; Lin, Y.-J.; Shao, Y.-H.; Chen, C.-A.; Chen, S.-L.; et al. A High-Accuracy Detection System: Based on Transfer Learning for Apical Lesions on Periapical Radiograph. Bioengineering 2022, 9, 777. https://doi.org/10.3390/bioengineering9120777
Chuo Y, Lin W-M, Chen T-Y, Chan M-L, Chang Y-S, Lin Y-R, Lin Y-J, Shao Y-H, Chen C-A, Chen S-L, et al. A High-Accuracy Detection System: Based on Transfer Learning for Apical Lesions on Periapical Radiograph. Bioengineering. 2022; 9(12):777. https://doi.org/10.3390/bioengineering9120777
Chicago/Turabian StyleChuo, Yueh, Wen-Ming Lin, Tsung-Yi Chen, Mei-Ling Chan, Yu-Sung Chang, Yan-Ru Lin, Yuan-Jin Lin, Yu-Han Shao, Chiung-An Chen, Shih-Lun Chen, and et al. 2022. "A High-Accuracy Detection System: Based on Transfer Learning for Apical Lesions on Periapical Radiograph" Bioengineering 9, no. 12: 777. https://doi.org/10.3390/bioengineering9120777